| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,670,381 – 3,670,487 |
| Length | 106 |
| Max. P | 0.723081 |

| Location | 3,670,381 – 3,670,487 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.99 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -4.96 |
| Energy contribution | -6.27 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.20 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3670381 106 + 22407834 AUUCCUGCACAUUGCUGUUUGAAAUAAAAUUUACCAUCAGUUAA-----------UA--AUGGAUAAUAAUAGUGUUUUGCUAACUACUUACGAUCUUUUUUCCCGUGAGCUAGUGCAA- .....(((((......(((.((((((....(((((((.......-----------..--)))).)))......))))))...)))..((((((...........))))))...))))).- ( -16.50) >DroSec_CAF1 132815 102 + 1 AUUCCUGCACAUUGCUGUUUGAAAGA-AAUUUAGCAGCAGUUAA-----------UA--AUGGUCA---AUGGUGGUUUACUAACUACUCACGAUCUUUUUGCCCUUGGGCAAGUGCAA- .....(((((((((((((...(((..-..))).))))))))...-----------..--..((((.---..(((((((....)))))))...))))...(((((....)))))))))).- ( -34.50) >DroSim_CAF1 138907 102 + 1 AUUCUUGCACAUUGCUGUUUGAAAGA-AAUUUAACAGCAGUUAA-----------UA--AUGGUCA---ACAGUGGUUUACUAACUACUUACGAUCUUUUUGCCCGUGAGCAAGUGCAA- .....((((((((((((((..(((..-..))))))))))))...-----------..--..((((.---..(((((((....)))))))...))))...((((......))))))))).- ( -29.40) >DroEre_CAF1 137602 91 + 1 ACU-UUGCACUUUGCUGUUCGAAAGA-AAUGUAUCAC----------------------UCAGUCA---AGAGUGAUUUCAUU-CUACUUACGAUCUCUUUUUCCCUGAACAAGUGCCA- ...-..(((((....((((((..(((-(.((.(((((----------------------((.....---.)))))))..))))-))......((........))..)))))))))))..- ( -21.50) >DroYak_CAF1 142518 102 + 1 UUUCUUGCACUUUGCUGUUUUAAAGA-AAUUUACCAAAAAUUAA-----------UA--ACAGUGA---GCAGUGGUUUCAUAACUACUUACGAUCAUUUUGUCCGUGAACAAGUGCAA- .....(((((((..(((((.((((..-..))))...........-----------.)--))))..)---...((((((....))))))(((((..(.....)..)))))...)))))).- ( -21.42) >DroAna_CAF1 148648 111 + 1 AUAACGACAGAUUGCUA-----GGGA-AAUUAAG---GAGUUAAAGGAGUAUAAAUAUUAUAAAUUAGUAUGGUAUUUUUUUAAUAACUUACGACUUUUUUGUGAGAAAGCAAGACCCAU .................-----(((.-...((((---..(((((((((((((..((((((.....)))))).)))))))))))))..))))......((((((......))))))))).. ( -23.40) >consensus AUUCCUGCACAUUGCUGUUUGAAAGA_AAUUUACCAGCAGUUAA___________UA__AUAGUCA___AUAGUGGUUUACUAACUACUUACGAUCUUUUUGCCCGUGAGCAAGUGCAA_ .....(((((.(((((.......................................................((((..........))))((((...........)))))))))))))).. ( -4.96 = -6.27 + 1.31)

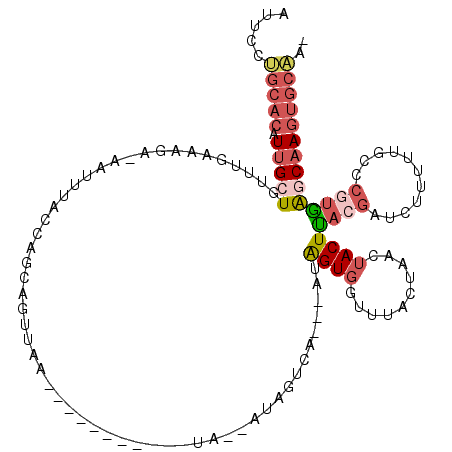

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:03 2006