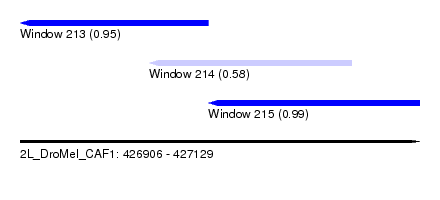
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 426,906 – 427,129 |
| Length | 223 |
| Max. P | 0.994311 |

| Location | 426,906 – 427,011 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.56 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -9.38 |
| Energy contribution | -10.35 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 426906 105 - 22407834 ACCGCAGG-------AAAAACACUUGUUCAAACAGAGGCGUGUGAAGUGUGCGGAAU---GUAUGCACAUAUACAAUUUCACACAUGACACAC-----AAAACAAUUUACUUAAAAUCAU .(((((((-------.......)))))((.....)))).(((((..(((((.(((((---(((((....))))).))))).)))))..)))))-----...................... ( -23.60) >DroSec_CAF1 4381 110 - 1 ACCGCAGG-------AAAAACACUUGUUCAAACAGAGGCGUGUGAAGUGUGCGGAAU---GUAUGCACAUAUACAAUUUCACACAUGACACACAUCUCAAAACAAUUUACUUAAAAUCAA ...(((((-------.......))))).......(((..(((((..(((((.(((((---(((((....))))).))))).)))))..)))))..)))...................... ( -28.30) >DroSim_CAF1 3366 110 - 1 ACCGCAGG-------AAAAACACUUGUUCAAACAGAGGCGUGUGAAGUGUGCGGAAU---GUAUGCACAUAUACAAUUUCACACAUGACACACAUCUCAAAACAAUUUACUUAAAAUCAA ...(((((-------.......))))).......(((..(((((..(((((.(((((---(((((....))))).))))).)))))..)))))..)))...................... ( -28.30) >DroEre_CAF1 3979 103 - 1 ACCGCAG--------G-AAACACUUGUUCAAACAGAGGCGUGUGAAGUGUGCGG-------AAUGUGCAUA-ACAAUUUCACACAUGACACACAUCUCAAAACAAUUUACUUUAAAUCAA ...((((--------(-.....))))).......(((..(((((..(((((.((-------(((((.....-)).))))).)))))..)))))..)))...................... ( -26.50) >DroYak_CAF1 3610 112 - 1 ACCGCAGG-------G-AAACACUUGUUCAAACAGAGGCGUGUGAAGUGUGCGGAACUUGGAAUGUACAUAUACAAUUUCACACAUGACACACAUCUCAAAACAAUUUACUUUAAAUCAA ...(((((-------.-.....))))).......(((..(((((..(((((.((((.(((.............))))))).)))))..)))))..)))...................... ( -26.52) >DroAna_CAF1 4243 110 - 1 ACCACAGGGACAAAAAAAAACACUUGUCAAAAUAACAGACUAUGGGAUAUGAGAGUU---AAAAAACAAUAUAGAGUUAAAGACUAA--AAAA-----AAAGGGGUUUAAAGAUAAUUAU .((.....(((((..........))))).........((((.....((((....(((---....))).))))..)))).........--....-----...))................. ( -7.50) >consensus ACCGCAGG_______AAAAACACUUGUUCAAACAGAGGCGUGUGAAGUGUGCGGAAU___GAAUGCACAUAUACAAUUUCACACAUGACACACAUCUCAAAACAAUUUACUUAAAAUCAA ...(((((..............)))))............((((((((((((((..........))))))).......))))))).................................... ( -9.38 = -10.35 + 0.98)

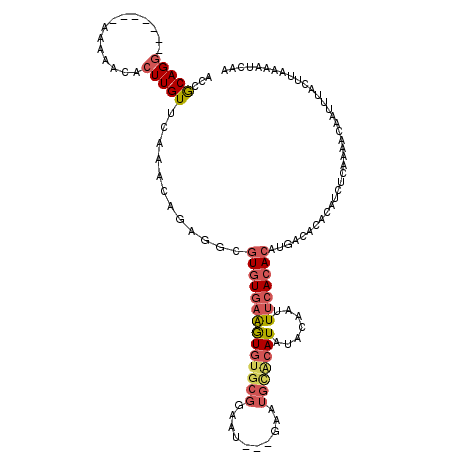

| Location | 426,978 – 427,091 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.19 |
| Mean single sequence MFE | -26.06 |
| Consensus MFE | -16.58 |
| Energy contribution | -17.17 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 426978 113 - 22407834 CGGCAAAGAGAAGGAGCGGGGGGGGGGGGCUGGUUGCCAUAUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAUAAACCGCAGG-------AAAAACACUUGUUCAAACAGAGGCG ..((.........(((((((.(.((((((((((((((((((....))))))))).))))))).........((....))..)).).(.-------.....).)))))))........)). ( -30.83) >DroSec_CAF1 4458 108 - 1 CGGGAAAGAGAAGGAGCUGG-----GGGGCUGGUUGCCAUAUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAUAAACCGCAGG-------AAAAACACUUGUUCAAACAGAGGCG .((............(((..-----((((((((((((((((....))))))))).))))))).))).....((....))..))(((((-------.......)))))............. ( -27.40) >DroSim_CAF1 3443 108 - 1 CGGGAAAGAGAAGGAGCUGG-----GGGGCUGGUUGCCAUAUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAGAAACCGCAGG-------AAAAACACUUGUUCAAACAGAGGCG .((............(((..-----((((((((((((((((....))))))))).))))))).)))(....).........))(((((-------.......)))))............. ( -26.00) >DroEre_CAF1 4051 106 - 1 CAGGAAAGAAGAGGAGCUCA-----GGGACUGGUUGCCAUAUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAUAAACCGCAG--------G-AAACACUUGUUCAAACAGAGGCG .......(((.(((.((.(.-----.(((((((((((((((....))))))))).))))))..)))....................(--------.-...).))).)))........... ( -29.20) >DroYak_CAF1 3690 107 - 1 CGGGAAGGAAAAGGAGCCGA-----GGGGCUGGUUGCCAUGUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAUAAACCGCAGG-------G-AAACACUUGUUCAAACAGAGGCG .((............(((..-----((((((((((((((((....))))))))).))))))).))).....((....))..))(((((-------.-.....)))))............. ( -30.20) >DroAna_CAF1 4313 106 - 1 -----AAGAUUGGGGGGUGG-----GGG-AGGGUAAU--GAUUAUUAUGGUAACACAGUUUUGGGCAAAAAUAGAAACA-ACCACAGGGACAAAAAAAAACACUUGUCAAAAUAACAGAC -----....((((.(((((.-----(..-(..(((((--....)))))..)..)...(((((.((..............-.))...))))).........))))).)))).......... ( -12.76) >consensus CGGGAAAGAGAAGGAGCUGG_____GGGGCUGGUUGCCAUAUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAUAAACCGCAGG_______AAAAACACUUGUUCAAACAGAGGCG .........(((.(((.........((((((((((((((((....))))))))).))))))).((................))...................))).)))........... (-16.58 = -17.17 + 0.59)


| Location | 427,011 – 427,129 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.26 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -27.96 |
| Energy contribution | -27.84 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 427011 118 - 22407834 CAAUGGAUUCACGGUAAUGCAAUUGGUUCCCAUGGGCUCGGCAAAGAGAAGGAGCGGGGGGGGGGGGCUGGUUGCCAUAUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAUAA .......(((.......(((......(((((....((((............))))...)))))((((((((((((((((....))))))))).)))))))..)))......))).... ( -32.12) >DroSec_CAF1 4491 113 - 1 CAAUGGAUUCACGGUAAUGUAUUUGGUUCCCAUGGGCUCGGGAAAGAGAAGGAGCUGG-----GGGGCUGGUUGCCAUAUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAUAA .......(((...........(((..(((((........)))))..)))....(((..-----((((((((((((((((....))))))))).))))))).))).))).......... ( -31.70) >DroSim_CAF1 3476 113 - 1 CAAUGGAUUCACGGUAAUGUAUUUGGUUCCCAUGGGCUCGGGAAAGAGAAGGAGCUGG-----GGGGCUGGUUGCCAUAUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAGAA .......(((...........(((..(((((........)))))..)))....(((..-----((((((((((((((((....))))))))).))))))).))).))).......... ( -31.70) >DroEre_CAF1 4082 113 - 1 CAAUGGAUGCACGGUAAUGCAUUUGGUUCCCAUGGGCUCAGGAAAGAAGAGGAGCUCA-----GGGACUGGUUGCCAUAUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAUAA ....(((((((......)))))))...((((.(((((((............)))))))-----))))((((((((((((....))))))))).)))(((((........))))).... ( -36.20) >DroYak_CAF1 3722 113 - 1 CAAUGGAUUCCCGGUAAUGCAUUUGGUUCCCAUGGGCUCGGGAAGGAAAAGGAGCCGA-----GGGGCUGGUUGCCAUGUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAUAA .......(((((((....((...(((...)))...))))))))).........(((..-----((((((((((((((((....))))))))).))))))).))).............. ( -36.60) >consensus CAAUGGAUUCACGGUAAUGCAUUUGGUUCCCAUGGGCUCGGGAAAGAGAAGGAGCUGG_____GGGGCUGGUUGCCAUAUUAUUAUGGUAACACAGUUUUAGGCAGAAAUAGAAAUAA .................(((..((((((((......(((......)))..)))))))).....((((((((((((((((....))))))))).)))))))..)))............. (-27.96 = -27.84 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:10 2006