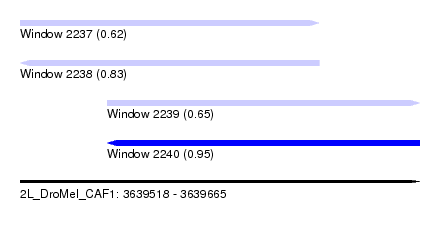
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,639,518 – 3,639,665 |
| Length | 147 |
| Max. P | 0.952065 |

| Location | 3,639,518 – 3,639,628 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -45.08 |
| Consensus MFE | -27.64 |
| Energy contribution | -28.68 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3639518 110 + 22407834 GGUGAUACA---GCACCGGACUUCCUUGGCUGGUGUACAGGCCCUGCGUCUGCGCCUGAAGCUGCUGCAAUUGCUGAUAGCGCUGCUGUAGCUGCAGCUCCUGUUGGUGCAGA ((.(.....---(((((((.((.....))))))))).....)))....((((((((...((..((((((...(((.(((((...))))))))))))))..))...)))))))) ( -44.80) >DroGri_CAF1 129137 113 + 1 GAUAGUGCACCGGCACUGGACUACCGGAUGCGGUAUAGAUGCCCUGUGUCUGUGCCUGCAACUGUUGCAAUUGCUGAUAGCGCUGCUGCAGCACCAGCUCCUGCUGAUGCAGC (((((((((...(((((((....)))).)))(((((((((((...)))))))))))))).))))))(((..(((((.((((...))))))))).((((....)))).)))... ( -46.50) >DroWil_CAF1 159504 110 + 1 GCUGGUAUA---GAACCGGACUCCCUGUGGCCGUAUAGAAACCUUGUGUCUGCGCCUGCAAUUGCUGCAAUUGUUGAUAGCGUUGCUGCAACUGCAGUUCCUGUUGAUGCAGC ((.((..((---((..(((.....(((((....))))).....)))..))))..)).))....((((((....(..((((.(..(((((....))))).)))))..))))))) ( -32.70) >DroYak_CAF1 112429 110 + 1 GAUGGUACA---GCACCGGACUGCCUUGGCUGGUGUACAGACCUUGGGUCUGUGCCUGCAGUUGUUGCAAUUGCUGAUAGCGCUGCUGCAGCUGCAGCUCCUGUUGGUGCAGU ..((((...---..)))).(((((.(..((.((.(((((((((...)))))))))((((((((((.(((..(((.....))).))).))))))))))..)).))..).))))) ( -49.90) >DroAna_CAF1 116752 110 + 1 GGUGGUACA---GCACCGGACUACCCUGGGCGGUGUACAGGCCCUGGGUCUGGGCCUGGAGCUGUUGCAGUUGCUGGUAGCGCUGCUGCAGUUGCAGCUCCUGUUGGUGCAAC .........---(((((((.((((((.((((.(....)..)))).))))..)).)).((((((((((((((.((.......)).))))))...))))))))....)))))... ( -51.50) >consensus GAUGGUACA___GCACCGGACUACCUUGGGCGGUGUACAGGCCCUGGGUCUGCGCCUGCAGCUGUUGCAAUUGCUGAUAGCGCUGCUGCAGCUGCAGCUCCUGUUGGUGCAGC ...(((......((((((............))))))....)))......(((((((.((((..((((((...((((.((((...))))))))))))))..)))).))))))). (-27.64 = -28.68 + 1.04)


| Location | 3,639,518 – 3,639,628 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -40.49 |
| Consensus MFE | -19.88 |
| Energy contribution | -21.76 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3639518 110 - 22407834 UCUGCACCAACAGGAGCUGCAGCUACAGCAGCGCUAUCAGCAAUUGCAGCAGCUUCAGGCGCAGACGCAGGGCCUGUACACCAGCCAAGGAAGUCCGGUGC---UGUAUCACC (((((.((....((((((((.(((.....)))((.....)).......)))))))).)).))))).((((.(((.(.((.((......))..))).))).)---)))...... ( -38.70) >DroGri_CAF1 129137 113 - 1 GCUGCAUCAGCAGGAGCUGGUGCUGCAGCAGCGCUAUCAGCAAUUGCAACAGUUGCAGGCACAGACACAGGGCAUCUAUACCGCAUCCGGUAGUCCAGUGCCGGUGCACUAUC ((.((((((((....)))))))).))....(((((....(((((((...))))))).(((((.(((...((....)).(((((....))))))))..))))))))))...... ( -46.30) >DroWil_CAF1 159504 110 - 1 GCUGCAUCAACAGGAACUGCAGUUGCAGCAACGCUAUCAACAAUUGCAGCAAUUGCAGGCGCAGACACAAGGUUUCUAUACGGCCACAGGGAGUCCGGUUC---UAUACCAGC .((((.((.....)).((((((((((.((((............)))).))))))))))..))))......(((.......(((((....))...)))....---...)))... ( -30.94) >DroYak_CAF1 112429 110 - 1 ACUGCACCAACAGGAGCUGCAGCUGCAGCAGCGCUAUCAGCAAUUGCAACAACUGCAGGCACAGACCCAAGGUCUGUACACCAGCCAAGGCAGUCCGGUGC---UGUACCAUC ((.(((((....((.(((((....))))).((((.....))..(((((.....)))))))(((((((...)))))))...)).((....)).....)))))---.))...... ( -40.60) >DroAna_CAF1 116752 110 - 1 GUUGCACCAACAGGAGCUGCAACUGCAGCAGCGCUACCAGCAACUGCAACAGCUCCAGGCCCAGACCCAGGGCCUGUACACCGCCCAGGGUAGUCCGGUGC---UGUACCACC ...(((((....(((((((....(((((..((.......))..))))).))))))).((.(...((((.((((.........)))).)))).).)))))))---......... ( -45.90) >consensus GCUGCACCAACAGGAGCUGCAGCUGCAGCAGCGCUAUCAGCAAUUGCAACAGCUGCAGGCACAGACACAGGGCCUGUACACCGCCCAAGGUAGUCCGGUGC___UGUACCACC ((((((.........(((((.......)))))((.....))...))))))...(((((((.(........))))))))..........((....))(((((....)))))... (-19.88 = -21.76 + 1.88)

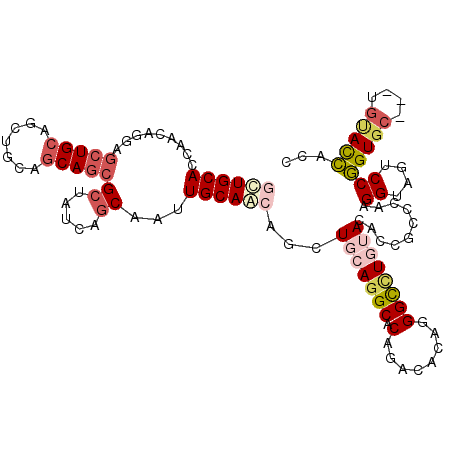
| Location | 3,639,550 – 3,639,665 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.03 |
| Mean single sequence MFE | -47.02 |
| Consensus MFE | -26.08 |
| Energy contribution | -26.84 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3639550 115 + 22407834 UACAGGCCCUGCGUCUGCGCCUGAAGCUGCUGCAAUUGCUGAUAGCGCUGCUGUAGCUGCAGCUCCUGUUGGUGCAGAAGGCUUCGGGGAUCCUGGUUCAAUUGCUG---GGCGCCCG ..((((((.....((((((((...((..((((((...(((.(((((...))))))))))))))..))...)))))))).))))).)(((..((..((......))..---))..))). ( -51.40) >DroGri_CAF1 129172 112 + 1 UAGAUGCCCUGUGUCUGUGCCUGCAACUGUUGCAAUUGCUGAUAGCGCUGCUGCAGCACCAGCUCCUGCUGAUGCAGCAGACUGCGUUGAUCUGUGUGC------UGUGGUGUACCCG .((((((...))))))(((((..((......(((...((.((((((((..((((.(((.((((....)))).))).))))...))))).))).)).)))------))..).))))... ( -39.30) >DroWil_CAF1 159536 118 + 1 UAGAAACCUUGUGUCUGCGCCUGCAAUUGCUGCAAUUGUUGAUAGCGUUGCUGCAACUGCAGUUCCUGUUGAUGCAGCAGACUCCUGGGAUCUGUGUGUAACUGCUGCAGCGUACCAC .(((..((..(.((((((..(((((.((((.(((((.((.....))))))).)))).)))))....(((....))))))))).)..))..)))(((.(((...((....)).)))))) ( -39.90) >DroYak_CAF1 112461 115 + 1 UACAGACCUUGGGUCUGUGCCUGCAGUUGUUGCAAUUGCUGAUAGCGCUGCUGCAGCUGCAGCUCCUGUUGGUGCAGUAGGCUGCGGGGAUCCUGGUUCAGUUGCUG---GGCGCCCG .(((((((...)))))))(((.((((..((((((...((((.((((...))))))))))))))..)))).)))((((....)))).(((..((..((......))..---))..))). ( -53.50) >DroAna_CAF1 116784 115 + 1 UACAGGCCCUGGGUCUGGGCCUGGAGCUGUUGCAGUUGCUGGUAGCGCUGCUGCAGUUGCAGCUCCUGUUGGUGCAACAAACUACGUGGAUCCUGGUGGAGUUGCUG---GGCACCCG ..(((((((.......))))))).((((((.(((((.((.....))))))).))))))(((((((((((((...))))).((((.(.....).)))))))))))).(---(....)). ( -51.00) >consensus UACAGGCCCUGGGUCUGCGCCUGCAGCUGUUGCAAUUGCUGAUAGCGCUGCUGCAGCUGCAGCUCCUGUUGGUGCAGCAGACUGCGGGGAUCCUGGUGCAAUUGCUG___GGCACCCG ..(((((.....)))))..(((((((((((((((...((((.((((...))))))))..((((....)))).)))))))).))))))).............................. (-26.08 = -26.84 + 0.76)

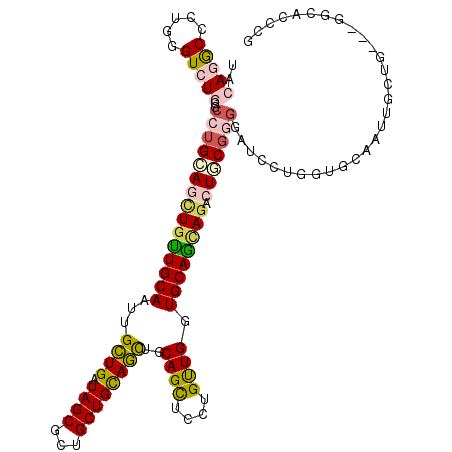
| Location | 3,639,550 – 3,639,665 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.03 |
| Mean single sequence MFE | -41.34 |
| Consensus MFE | -18.36 |
| Energy contribution | -19.96 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3639550 115 - 22407834 CGGGCGCC---CAGCAAUUGAACCAGGAUCCCCGAAGCCUUCUGCACCAACAGGAGCUGCAGCUACAGCAGCGCUAUCAGCAAUUGCAGCAGCUUCAGGCGCAGACGCAGGGCCUGUA (((((.((---..((..(((..((.((....))(((((.(((((......)))))(((((((((.....)))((.....))...)))))).))))).))..)))..)).))))))).. ( -41.80) >DroGri_CAF1 129172 112 - 1 CGGGUACACCACA------GCACACAGAUCAACGCAGUCUGCUGCAUCAGCAGGAGCUGGUGCUGCAGCAGCGCUAUCAGCAAUUGCAACAGUUGCAGGCACAGACACAGGGCAUCUA .((.....))...------((.....(((...(((.(.((((.((((((((....)))))))).))))).)))..))).(((((((...)))))))..)).................. ( -37.00) >DroWil_CAF1 159536 118 - 1 GUGGUACGCUGCAGCAGUUACACACAGAUCCCAGGAGUCUGCUGCAUCAACAGGAACUGCAGUUGCAGCAACGCUAUCAACAAUUGCAGCAAUUGCAGGCGCAGACACAAGGUUUCUA ((((((.((((...))))))).)))(((..((..(.((((((.((.((.....)).((((((((((.((((............)))).)))))))))))))))))).)..))..))). ( -43.30) >DroYak_CAF1 112461 115 - 1 CGGGCGCC---CAGCAACUGAACCAGGAUCCCCGCAGCCUACUGCACCAACAGGAGCUGCAGCUGCAGCAGCGCUAUCAGCAAUUGCAACAACUGCAGGCACAGACCCAAGGUCUGUA ..(((((.---......(((.....((....))((((....)))).....)))..(((((....))))).)))))....((...((((.....)))).))(((((((...))))))). ( -41.20) >DroAna_CAF1 116784 115 - 1 CGGGUGCC---CAGCAACUCCACCAGGAUCCACGUAGUUUGUUGCACCAACAGGAGCUGCAACUGCAGCAGCGCUACCAGCAACUGCAACAGCUCCAGGCCCAGACCCAGGGCCUGUA ..(((((.---.(((((((((....)))........).))))))))))....(((((((....(((((..((.......))..))))).)))))))((((((.......))))))... ( -43.40) >consensus CGGGUGCC___CAGCAACUGCACCAGGAUCCCCGCAGUCUGCUGCACCAACAGGAGCUGCAGCUGCAGCAGCGCUAUCAGCAAUUGCAACAGCUGCAGGCACAGACACAGGGCCUGUA .................................((((.((((((((.........(((((.......)))))((.....))...))))))))))))((((.(........)))))... (-18.36 = -19.96 + 1.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:52 2006