
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,629,222 – 3,629,325 |
| Length | 103 |
| Max. P | 0.970845 |

| Location | 3,629,222 – 3,629,325 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 104 |
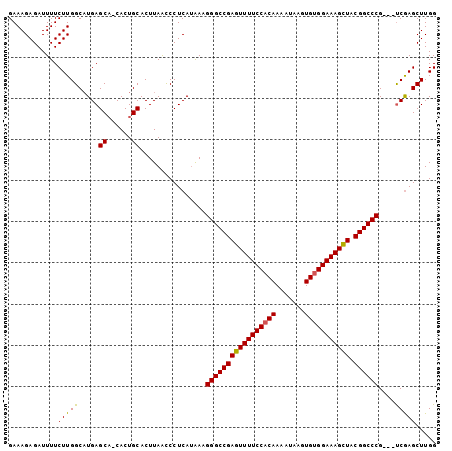
| Reading direction | forward |
| Mean pairwise identity | 90.26 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -32.92 |
| Energy contribution | -32.37 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
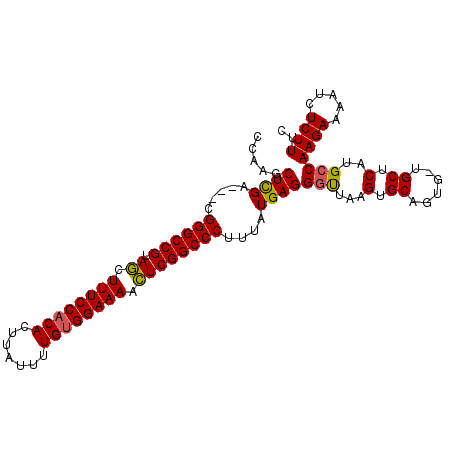
>2L_DroMel_CAF1 3629222 103 + 22407834 GAAAGAGAUUUUCUUGGCAUGAGCA-CACUGCACUUAGCCCUCAUAAAGGGCCGAAUUUUCCUCAAAAUAAGUGUGGAAAGUUACGGCCCGCGGCCAAGCUUGG ............(((((((((((..-..(((....)))..)))))...((((((((((((((.((.......)).)))))))).))))))...))))))..... ( -35.70) >DroSec_CAF1 97042 101 + 1 GAAAGAGAUUUUCUUGGCAUGAGCUCCACUGCACUUAAUCCUCACAAAGGGCCGAGUUUUCCACAAAAUAAGUGUGGAAAGCUACGGCCCG---UCGAGCUUGG ............((((((....((......))................(((((((((((((((((.......))))))))))).)))))))---)))))..... ( -38.30) >DroSim_CAF1 102320 100 + 1 GAAAGAGAUUUUCUUGGCUUGAGCA-CACUGCACUUAACCCUCAUGAAGGGCCGAGUUUUCCACAAAAUAAGUGUGGAAAGCUACGGCCCG---UCGAGCUUGG ............((.(((((((((.-....))................(((((((((((((((((.......))))))))))).)))))).---))))))).)) ( -40.30) >consensus GAAAGAGAUUUUCUUGGCAUGAGCA_CACUGCACUUAACCCUCAUAAAGGGCCGAGUUUUCCACAAAAUAAGUGUGGAAAGCUACGGCCCG___UCGAGCUUGG ............(((((.....((......))................(((((((((((((((((.......))))))))))).))))))....)))))..... (-32.92 = -32.37 + -0.55)


| Location | 3,629,222 – 3,629,325 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 104 |
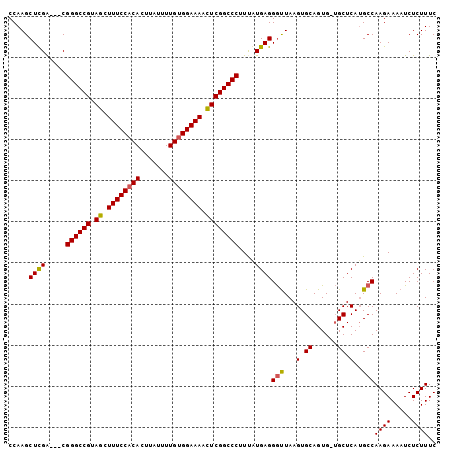
| Reading direction | reverse |
| Mean pairwise identity | 90.26 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -29.05 |
| Energy contribution | -28.83 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3629222 103 - 22407834 CCAAGCUUGGCCGCGGGCCGUAACUUUCCACACUUAUUUUGAGGAAAAUUCGGCCCUUUAUGAGGGCUAAGUGCAGUG-UGCUCAUGCCAAGAAAAUCUCUUUC .....((((((...((((((.((.(((((.((.......)).))))).))))))))...(((((.((..........)-).)))))))))))............ ( -34.90) >DroSec_CAF1 97042 101 - 1 CCAAGCUCGA---CGGGCCGUAGCUUUCCACACUUAUUUUGUGGAAAACUCGGCCCUUUGUGAGGAUUAAGUGCAGUGGAGCUCAUGCCAAGAAAAUCUCUUUC ...(((((.(---(((((((.((.((((((((.......)))))))).))))))))..(((...........))))).)))))......((((.....)))).. ( -34.80) >DroSim_CAF1 102320 100 - 1 CCAAGCUCGA---CGGGCCGUAGCUUUCCACACUUAUUUUGUGGAAAACUCGGCCCUUCAUGAGGGUUAAGUGCAGUG-UGCUCAAGCCAAGAAAAUCUCUUUC .....(((((---.((((((.((.((((((((.......)))))))).)))))))).))..)))((((.((..(...)-..))..))))((((.....)))).. ( -34.00) >consensus CCAAGCUCGA___CGGGCCGUAGCUUUCCACACUUAUUUUGUGGAAAACUCGGCCCUUUAUGAGGGUUAAGUGCAGUG_UGCUCAUGCCAAGAAAAUCUCUUUC .....((((.....((((((.((.((((((((.......)))))))).))))))))....))))(((...(.((......)).)..)))((((.....)))).. (-29.05 = -28.83 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:47 2006