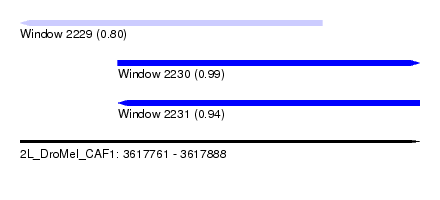
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,617,761 – 3,617,888 |
| Length | 127 |
| Max. P | 0.986767 |

| Location | 3,617,761 – 3,617,857 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.21 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -24.31 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3617761 96 - 22407834 GCUCGA-CACCAGCUAAUGGGAGUGGGCGUUGUGUCGACCCACUUCCCAGGCCAUUGCGAUGCCGAUUGGCUCUGG-GCGAAACACU--CUCACACACUU ..(((.-..((((....((((((((((((......)).))))).)))))(((((...((....))..)))))))))-.)))......--........... ( -35.00) >DroSec_CAF1 85504 96 - 1 GCUCCA-CACCAGCUAAUGGGAGUGGGCGUUGUGUCGACCCACUUCCCAGGCAAUUUCGAUGCCGAUUGGCUCUGG-GCGAAACACU--CUCUCACACUU ((.(((-....((((((((((((((((((......)).)))))))))..((((.......)))).))))))).)))-))........--........... ( -35.00) >DroSim_CAF1 90860 96 - 1 GCUCCA-CACCAGCUAAUGGGAGUGGGCGUUGUGUCGACCCACUUCCCAGGCCAUUUCGAUGCCGAUUGGCUCUGG-GCGAAACACU--CUCUCACACUU ......-.....(((...(((((((((((......)).)))))))))(((((((..(((....))).)))).))))-))........--........... ( -33.50) >DroEre_CAF1 86967 87 - 1 GCCCCACCACCAGCUGAUGGGAGUGGGCGUAGUAUCGACCCACUUCCCAGGCCAUUUCGAUGCCGAUUGGCUUUGG-------------CUCUCACACUU ...........(((..(((((((((((((......)).))))).)))))(((((..(((....))).))))))..)-------------))......... ( -32.40) >DroYak_CAF1 90057 99 - 1 GCUCCA-CACCAGCUAAUGGGAGUGGGCGUUGUAUCGACCCACUUCCCAGGCCAUUGCGAUGCCGAUUGGCUCUGGGGCGAAUCACUCUCUCCCACACUU ((((((-....((((((((((((((((((......)).)))))))))..(((.........))).))))))).))))))..................... ( -37.00) >DroAna_CAF1 92684 84 - 1 --------------UGUGGGGCGUGGGCGUUCUAUCGACCCACUUCCCAGGCCCUGGCACUGCCGAUUGGCUCUUG-G-AAAACACUCACUCUCACACUC --------------(((((((.(((((((......)).)))))((((.(((((.((((...))))...))).)).)-)-)).........)))))))... ( -30.70) >consensus GCUCCA_CACCAGCUAAUGGGAGUGGGCGUUGUAUCGACCCACUUCCCAGGCCAUUGCGAUGCCGAUUGGCUCUGG_GCGAAACACU__CUCUCACACUU ..................(((((((((((......)).)))))))))(((((((...((....))..)))).)))......................... (-24.31 = -25.00 + 0.69)

| Location | 3,617,792 – 3,617,888 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -26.74 |
| Energy contribution | -27.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3617792 96 + 22407834 UCGGCAUCGCAAUGGCCUGGGAAGUGGGUCGACACAACGCCCACUCCCAUUAGCUGGUG-UCGAGCCACUCCAACG-CUCAACUUCCCCA-------CCACCCAC ..(((.(((((..(((.(((((.(((((.((......))))))))))))...)))..))-.)))))).........-.............-------........ ( -31.80) >DroSec_CAF1 85535 96 + 1 UCGGCAUCGAAAUUGCCUGGGAAGUGGGUCGACACAACGCCCACUCCCAUUAGCUGGUG-UGGAGCCACUCCAGCG-CUCAACUCCCCCA-------CCACCCAC ..((((.......))))(((((.(((((.((......))))))))))))...(((((.(-((....))).))))).-.............-------........ ( -38.40) >DroSim_CAF1 90891 96 + 1 UCGGCAUCGAAAUGGCCUGGGAAGUGGGUCGACACAACGCCCACUCCCAUUAGCUGGUG-UGGAGCCACUCCAACG-CUCAACUCCCCCA-------CCACCCAC ..(((.........)))(((((.(((((.((......)))))))))))).....(((((-.((((...........-.....))))..))-------)))..... ( -30.89) >DroEre_CAF1 86988 82 + 1 UCGGCAUCGAAAUGGCCUGGGAAGUGGGUCGAUACUACGCCCACUCCCAUCAGCUGGUGGUGGGGCCACUCCAACG-----------------------CCCUAC ..(((.......((((((((((.(((((.((......)))))))))))..((.(....).)))))))).......)-----------------------)).... ( -32.94) >DroYak_CAF1 90091 104 + 1 UCGGCAUCGCAAUGGCCUGGGAAGUGGGUCGAUACAACGCCCACUCCCAUUAGCUGGUG-UGGAGCCACUCCAACUACUCUACUCCCCCCCUCCCAGCCACCCAC ..(((........(((.(((((.(((((.((......))))))))))))...)))((.(-(((((............)))))).))..........)))...... ( -35.10) >consensus UCGGCAUCGAAAUGGCCUGGGAAGUGGGUCGACACAACGCCCACUCCCAUUAGCUGGUG_UGGAGCCACUCCAACG_CUCAACUCCCCCA_______CCACCCAC ..(((.(((...((((.(((((.(((((.((......))))))))))))...))))....))).)))...................................... (-26.74 = -27.14 + 0.40)

| Location | 3,617,792 – 3,617,888 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -42.80 |
| Consensus MFE | -30.03 |
| Energy contribution | -30.87 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3617792 96 - 22407834 GUGGGUGG-------UGGGGAAGUUGAG-CGUUGGAGUGGCUCGA-CACCAGCUAAUGGGAGUGGGCGUUGUGUCGACCCACUUCCCAGGCCAUUGCGAUGCCGA ....(..(-------(((((..((((((-(.........))))))-).))......((((((((((((......)).))))).)))))..))))..)........ ( -39.00) >DroSec_CAF1 85535 96 - 1 GUGGGUGG-------UGGGGGAGUUGAG-CGCUGGAGUGGCUCCA-CACCAGCUAAUGGGAGUGGGCGUUGUGUCGACCCACUUCCCAGGCAAUUUCGAUGCCGA .(((.(((-------((..(((((((..-(....)..))))))).-))))).))).((((((((((((......)).))))).)))))((((.......)))).. ( -45.00) >DroSim_CAF1 90891 96 - 1 GUGGGUGG-------UGGGGGAGUUGAG-CGUUGGAGUGGCUCCA-CACCAGCUAAUGGGAGUGGGCGUUGUGUCGACCCACUUCCCAGGCCAUUUCGAUGCCGA .(((.(((-------((..(((((((..-(....)..))))))).-))))).))).((((((((((((......)).))))).)))))(((.........))).. ( -42.90) >DroEre_CAF1 86988 82 - 1 GUAGGG-----------------------CGUUGGAGUGGCCCCACCACCAGCUGAUGGGAGUGGGCGUAGUAUCGACCCACUUCCCAGGCCAUUUCGAUGCCGA ....((-----------------------(((((((((((((.((.(....).)).((((((((((((......)).))))).)))))))))))))))))))).. ( -44.50) >DroYak_CAF1 90091 104 - 1 GUGGGUGGCUGGGAGGGGGGGAGUAGAGUAGUUGGAGUGGCUCCA-CACCAGCUAAUGGGAGUGGGCGUUGUAUCGACCCACUUCCCAGGCCAUUGCGAUGCCGA (..(.(((((..................(((((((.(((....))-).))))))).((((((((((((......)).))))).)))))))))))..)........ ( -42.60) >consensus GUGGGUGG_______UGGGGGAGUUGAG_CGUUGGAGUGGCUCCA_CACCAGCUAAUGGGAGUGGGCGUUGUGUCGACCCACUUCCCAGGCCAUUUCGAUGCCGA .............................(((((((((((((..............((((((((((((......)).))))).)))))))))))))))))).... (-30.03 = -30.87 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:43 2006