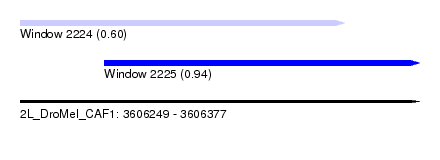
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,606,249 – 3,606,377 |
| Length | 128 |
| Max. P | 0.943365 |

| Location | 3,606,249 – 3,606,353 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.88 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -19.58 |
| Energy contribution | -18.70 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
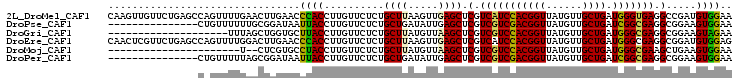
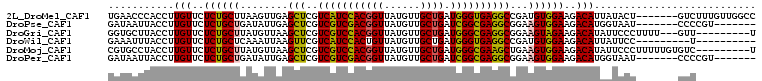
>2L_DroMel_CAF1 3606249 104 + 22407834 CAAGUUGUUCUGAGCCAGUUUUGAACUUGAACCCACCUUGUUCUCUGCUUAAGUUGAGCUCGUCAUCCACGGUUAUGUUGCUGAUGGGUGAGGCCGAUGUGGAA ((((((...(((...))).....))))))...((((.(((..(...((((.....))))...(((((((((((......)))).))))))))..))).)))).. ( -29.40) >DroPse_CAF1 81398 89 + 1 ---------------CUGUUUUUUGCGGAUAAUUACCUUGUUCUCUGCUGAUAUUGAGCUCGUCGUCGACGGUUAUGUUGCUGAUCGGCGAGGCGGAAGUGGAA ---------------(..(((((.(((((.(((......))).))))).........(((..(((((((((((......)))).)))))))))))))))..).. ( -26.30) >DroGri_CAF1 89123 84 + 1 --------------------UUUAGCUGGUGCUUACCUUGUUCUCUGCUUAUGUUAAGCUCGUCGUCCACGGUUAUGUUGCUGAUGGGCGAGGCGGAAGUAGAA --------------------...(((.(((....)))..))).(((((((.......(((..(((((((((((......)))).))))))))))..))))))). ( -25.50) >DroEre_CAF1 75449 104 + 1 CAACUCGUUCUGAGCCAGUUUUGGACUUGAACCCACCUUGUUCUCUGCUUAAGUUGAGCUCGUCAUCCACGGUUAUGUUGCUGAUGGGCGAGGCGGAUGUGGAG ...(((((((.(((((......)).)))))))((.((((....((.((....)).))((((((((.(.(((....))).).)))))))))))).)).....))) ( -29.90) >DroMoj_CAF1 96774 80 + 1 ----------------------U--CUCGUGCCUACCUUGUUCUCUGCUUAUGUUAAGCUCGUCGUCCACGGUUAUGUUGCUGAUGGGCGAAGCUGAAGUGGAA ----------------------.--..................((..(((......((((..(((((((((((......)))).))))))))))).)))..)). ( -24.10) >DroPer_CAF1 81677 89 + 1 ---------------CUGUUUUUAGCGGAUAAUUACCUUGUUCUCUGCUGAUAUUGAGCUCGUCGUCGACGGUUAUGUUGCUGAUCGGCGAGGCGGAAGUGGAA ---------------(..(((((((((((.(((......))).))))))).......(((..(((((((((((......)))).)))))))))).))))..).. ( -28.30) >consensus _______________C_GUUUUUAGCUGGUACCUACCUUGUUCUCUGCUUAUGUUGAGCUCGUCGUCCACGGUUAUGUUGCUGAUGGGCGAGGCGGAAGUGGAA ................................((((..........((((.....)))).(.(((((((((((......)))).))))))).).....)))).. (-19.58 = -18.70 + -0.88)
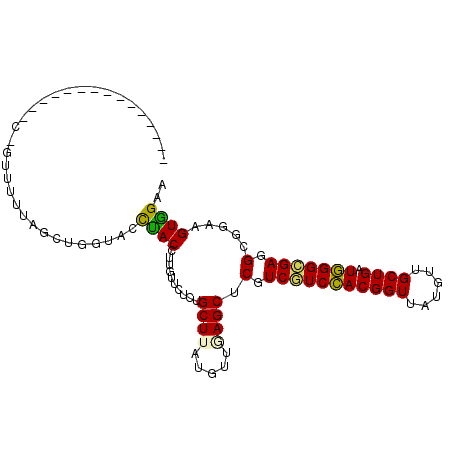
| Location | 3,606,276 – 3,606,377 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.44 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -23.00 |
| Energy contribution | -22.20 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3606276 101 + 22407834 UGAACCCACCUUGUUCUCUGCUUAAGUUGAGCUCGUCAUCCACGGUUAUGUUGCUGAUGGGUGAGGCCGAUGUGGAAGACAUUAUACU-------GUCUUUGUUGGCC .((((.......))))...((((.....))))...(((((((((((......)))).)))))))(((((((...(((((((......)-------))))))))))))) ( -33.60) >DroPse_CAF1 81410 94 + 1 GAUAAUUACCUUGUUCUCUGCUGAUAUUGAGCUCGUCGUCGACGGUUAUGUUGCUGAUCGGCGAGGCGGAAGUGGAAGACAUGGUAAU-------CCCCGU------- ....((((((.(((..((..((........(((..(((((((((((......)))).))))))))))...))..))..))).))))))-------......------- ( -32.50) >DroGri_CAF1 89130 96 + 1 GGUGCUUACCUUGUUCUCUGCUUAUGUUAAGCUCGUCGUCCACGGUUAUGUUGCUGAUGGGCGAGGCGGAAGUAGAAGACAUAUUCCCUUUU---GUU---------U (((....))).(((..(((((((.......(((..(((((((((((......)))).))))))))))..)))))))..)))...........---...---------. ( -29.40) >DroWil_CAF1 106719 89 + 1 GAAAUUUACCUUGUUCUCUGCUCAAAUUAAGUUCGUCAUCCACUGUUAUGUUGCUGAUGGGUGAGGCCGAUGUGGAAGACAUUAUUCC---------U---------- ...........(((..((..(((..........(((((.(.((......)).).)))))((.....)))).)..))..))).......---------.---------- ( -16.60) >DroMoj_CAF1 96777 99 + 1 CGUGCCUACCUUGUUCUCUGCUUAUGUUAAGCUCGUCGUCCACGGUUAUGUUGCUGAUGGGCGAAGCUGAAGUGGAAGACAUAUUCCCUUUUUGUGUC---------U ................((..(((......((((..(((((((((((......)))).))))))))))).)))..))(((((((.........))))))---------) ( -29.80) >DroPer_CAF1 81689 94 + 1 GAUAAUUACCUUGUUCUCUGCUGAUAUUGAGCUCGUCGUCGACGGUUAUGUUGCUGAUCGGCGAGGCGGAAGUGGAAGACAUGGUAAU-------CCCCGU------- ....((((((.(((..((..((........(((..(((((((((((......)))).))))))))))...))..))..))).))))))-------......------- ( -32.50) >consensus GAUACUUACCUUGUUCUCUGCUUAUAUUAAGCUCGUCGUCCACGGUUAUGUUGCUGAUGGGCGAGGCGGAAGUGGAAGACAUAAUACC_______GUC__________ ...........(((..((((((........(((..(((((((((((......)))).))))))))))...))))))..)))........................... (-23.00 = -22.20 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:38 2006