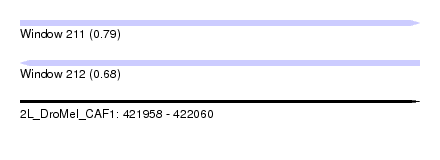
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 421,958 – 422,060 |
| Length | 102 |
| Max. P | 0.785840 |

| Location | 421,958 – 422,060 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -18.82 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 421958 102 + 22407834 GAUAAAAAUAAGCCUCCUCCC---------UCAGAAGCUCCAGCUAACAUUCCUCAUGGCGGUAAAAAUCUGCCCAAGUCCAUACAGAUCCUGGAAAUUGGAGGGAGUAUG ................(((((---------((..((..(((((..............(((((.......)))))...(((......))).)))))..)).))))))).... ( -26.30) >DroSec_CAF1 1883 102 + 1 GAUAAAAAUAAGCCCCCUCCC---------GCAGAAGCCCCAGCUAACAUUCCUCAUGGCGGUAAAAAUCUGCCCAAAUCCAUACAGAUCCUGGAAAUUGGAGGGAGUAUG ...........((.((((((.---------(((((.((((((..............))).))).....)))))(((.(((......)))..))).....)))))).))... ( -26.64) >DroSim_CAF1 1876 102 + 1 GAUAAAAAUAAGCCCCCUCCC---------GCUGAAACUCCAGCUAACAUUCCUCACGGCGGUAAAAAUCUGCCCAAAUCCAUACAGAUCCUGGAAAUUGGAGGGAGUAUG ...........((.((((((.---------((((......)))).....((((....(((((.......)))))...(((......)))...))))...)))))).))... ( -28.20) >DroEre_CAF1 1825 102 + 1 GAUAAACACAAGCCGCCUCCG---------GCAGAAACUCAAGCUAACAUUCCGCAUGGCGGGAAGAAUCUGCCCAAAUCCAUACAGAUCCUGGAAAUUGGAGGGAGCAUG ...........((..((((((---------(((((..((..........((((((...))))))))..))))))....((((.........))))....)))))..))... ( -29.60) >DroYak_CAF1 1862 102 + 1 GAUAAAAACAAGCCCCCUCCG---------AAAGAAGCUCCAGCUAACAUUCCGCACGGCGGUAAAAAUCUGCCCAAAUCCAUACAGAUCAUGGAAAUUGGAGGGAGUAUG ........((.((.(((((((---------(....(((....)))............(((((.......)))))....(((((.......)))))..)))))))).)).)) ( -28.70) >DroMoj_CAF1 1751 108 + 1 GACAGGCACAAACCACCACCGCCGCCAACAACAGAUG---CCGUAGCCACACCGCAUGGCGGCAAACACCUGCCUAAGUUUAUACAGGUGCUGGAAAUUGGAGGCAGCAUG ....((......))......(((.((((...(((.((---((((.((......))...))))))..((((((............)))))))))....)))).)))...... ( -32.60) >consensus GAUAAAAACAAGCCCCCUCCC_________GCAGAAGCUCCAGCUAACAUUCCGCAUGGCGGUAAAAAUCUGCCCAAAUCCAUACAGAUCCUGGAAAUUGGAGGGAGUAUG ...........((.((((((.....................................(((((.......)))))....((((.........))))....)))))).))... (-18.82 = -19.02 + 0.20)

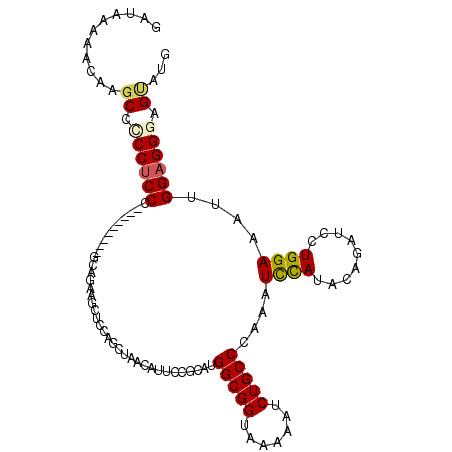
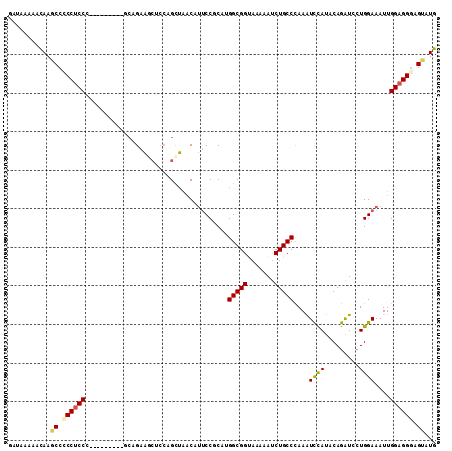
| Location | 421,958 – 422,060 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -22.21 |
| Energy contribution | -22.05 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 421958 102 - 22407834 CAUACUCCCUCCAAUUUCCAGGAUCUGUAUGGACUUGGGCAGAUUUUUACCGCCAUGAGGAAUGUUAGCUGGAGCUUCUGA---------GGGAGGAGGCUUAUUUUUAUC ....(((((((....(((((((((((((..........)))))))....((.......))........)))))).....))---------)))))................ ( -28.00) >DroSec_CAF1 1883 102 - 1 CAUACUCCCUCCAAUUUCCAGGAUCUGUAUGGAUUUGGGCAGAUUUUUACCGCCAUGAGGAAUGUUAGCUGGGGCUUCUGC---------GGGAGGGGGCUUAUUUUUAUC ....((((((((.....((((.((((....))))))))(((((......(((((((.....)))...)).))....)))))---------.))))))))............ ( -32.00) >DroSim_CAF1 1876 102 - 1 CAUACUCCCUCCAAUUUCCAGGAUCUGUAUGGAUUUGGGCAGAUUUUUACCGCCGUGAGGAAUGUUAGCUGGAGUUUCAGC---------GGGAGGGGGCUUAUUUUUAUC ....(((((((((((((((..(((((....)))))..)).))))).......((....)).......(((((....)))))---------.))))))))............ ( -33.20) >DroEre_CAF1 1825 102 - 1 CAUGCUCCCUCCAAUUUCCAGGAUCUGUAUGGAUUUGGGCAGAUUCUUCCCGCCAUGCGGAAUGUUAGCUUGAGUUUCUGC---------CGGAGGCGGCUUGUGUUUAUC ((.(((.(((((........((((((....)))))).((((((..(((.((((...))))...(....)..)))..)))))---------)))))).))).))........ ( -33.70) >DroYak_CAF1 1862 102 - 1 CAUACUCCCUCCAAUUUCCAUGAUCUGUAUGGAUUUGGGCAGAUUUUUACCGCCGUGCGGAAUGUUAGCUGGAGCUUCUUU---------CGGAGGGGGCUUGUUUUUAUC ((..((((((((....((((((.....))))))(((.(((.(((..((.((((...)))))).))).))).))).......---------.))))))))..))........ ( -32.10) >DroMoj_CAF1 1751 108 - 1 CAUGCUGCCUCCAAUUUCCAGCACCUGUAUAAACUUAGGCAGGUGUUUGCCGCCAUGCGGUGUGGCUACGG---CAUCUGUUGUUGGCGGCGGUGGUGGUUUGUGCCUGUC ((.((..((.((.......(((((((((.((....)).))))))))).(((((((.((((....((....)---)..))))...))))))))).))..)).))........ ( -47.60) >consensus CAUACUCCCUCCAAUUUCCAGGAUCUGUAUGGAUUUGGGCAGAUUUUUACCGCCAUGAGGAAUGUUAGCUGGAGCUUCUGC_________CGGAGGGGGCUUAUUUUUAUC ....((((((((.......(((((((((..........))))))))).....((....))...............................))))))))............ (-22.21 = -22.05 + -0.16)
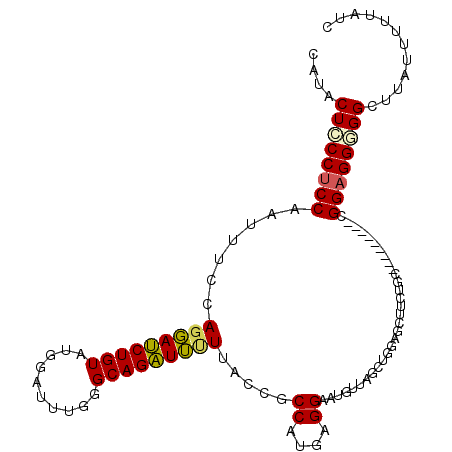
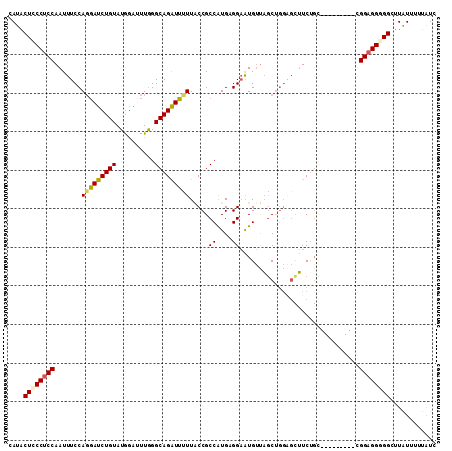
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:07 2006