
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,594,367 – 3,594,514 |
| Length | 147 |
| Max. P | 0.999048 |

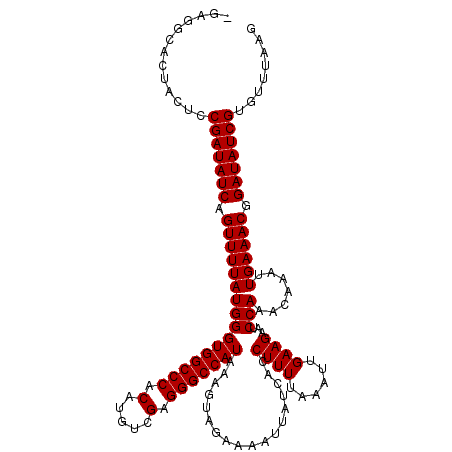
| Location | 3,594,367 – 3,594,486 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.60 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -28.11 |
| Energy contribution | -28.11 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3594367 119 + 22407834 -CAGGCACUACUCCGAUAUCAGUUUUAUGGGUGGCCCACAUGUCGAGGGCCAUAAAGUAGAAAAUUAUCACCUUUUAAAUUGAAGAAUCCAAACAAAUUGAAACGGAUAUCGUGUUUAAG -.((((((...((((...(((((((..((((((((((.(.....).)))))))..................((((......))))...)))...)))))))..))))....))))))... ( -30.90) >DroVir_CAF1 74863 112 + 1 --------UACUCCGAUAUCAGUUUUAUGGGUGGCCCACAUGUCGAGGGCCAUAAAGUAGAAAAUUAUCACCUUUUAAAUUGAAGAAUCCAAACAAAUUGAAACGGAUAUCGUGUUUAAG --------.....(((((((.((((((((((((((((.(.....).)))))))..................((((......))))...))).......)))))).)))))))........ ( -28.11) >DroGri_CAF1 76511 120 + 1 CGAGGCACUACUCCGAUAUCAGUUUUAUGGGUGGCCCACAUGUCGAGGGCCAUAAAGUAGAAAAUUAUCACCUUUUAAAUUGAAGAAUCCAAACAAAUUGAAACGGAUAUCGUGUUUAAG ..((((((...((((...(((((((..((((((((((.(.....).)))))))..................((((......))))...)))...)))))))..))))....))))))... ( -31.00) >DroEre_CAF1 63888 119 + 1 -CAGGCACUACUCCGAUAUCAGUUUUAUGGGUGGCCCACAUGUCGAGGGCCAUAAAGUAGAAAAUUAUCACCUUUUAAAUUGAAGAAUCCAAACAAAUUGAAACGGAUAUCGUGUUUAAG -.((((((...((((...(((((((..((((((((((.(.....).)))))))..................((((......))))...)))...)))))))..))))....))))))... ( -30.90) >DroWil_CAF1 93269 119 + 1 -GUGGCACUACUCCGAUAUCAGUUUUAUGGGUGGCCCACAUGUCGUGGGCCAUAAAGUAGAAAAUUAUCACCUUUUAAAUUGAAGAAUCCAAACAAAUUGAAACGGAUAUCGUGUUUAAG -..(((((...((((...(((((((..((((((((((((.....)))))))))..................((((......))))...)))...)))))))..))))....))))).... ( -34.30) >DroMoj_CAF1 83129 120 + 1 CGGGGCACUACUCCGAUAUCAGUUUUAUGGGUGGCCCACAUGUCGAGGGCCAUAAAGUAGAAAAUUAUCACCUUUUAAAUUGAAGAAUCCAAACAAAUUGAAACGGAUAUCGUGUUUAAG .((((.....))))((((((.((((((((((((((((.(.....).)))))))..................((((......))))...))).......)))))).))))))......... ( -31.21) >consensus _GAGGCACUACUCCGAUAUCAGUUUUAUGGGUGGCCCACAUGUCGAGGGCCAUAAAGUAGAAAAUUAUCACCUUUUAAAUUGAAGAAUCCAAACAAAUUGAAACGGAUAUCGUGUUUAAG .............(((((((.((((((((((((((((.(.....).)))))))..................((((......))))...))).......)))))).)))))))........ (-28.11 = -28.11 + -0.00)

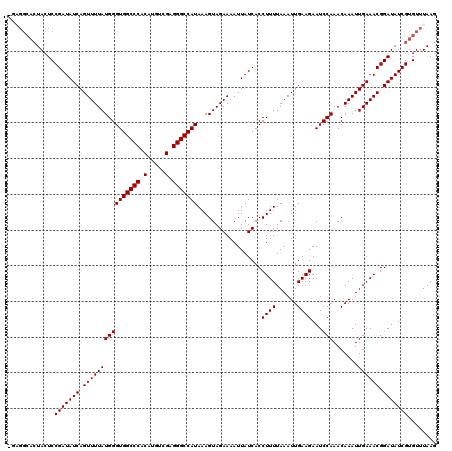
| Location | 3,594,367 – 3,594,486 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.60 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -27.88 |
| Energy contribution | -28.38 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3594367 119 - 22407834 CUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACAUGUGGGCCACCCAUAAAACUGAUAUCGGAGUAGUGCCUG- ......(((....((((..(((.(((...(((..............(((((((......)).))))).((((((.(.....).)))))).)))..))).)))...))))...)))....- ( -29.60) >DroVir_CAF1 74863 112 - 1 CUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACAUGUGGGCCACCCAUAAAACUGAUAUCGGAGUA-------- .....((.(((((((.((((.........(((..............(((((((......)).))))).((((((.(.....).)))))).)))..)))).)))))))..)).-------- ( -26.30) >DroGri_CAF1 76511 120 - 1 CUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACAUGUGGGCCACCCAUAAAACUGAUAUCGGAGUAGUGCCUCG ......(((....((((..(((.(((...(((..............(((((((......)).))))).((((((.(.....).)))))).)))..))).)))...))))...)))..... ( -29.60) >DroEre_CAF1 63888 119 - 1 CUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACAUGUGGGCCACCCAUAAAACUGAUAUCGGAGUAGUGCCUG- ......(((....((((..(((.(((...(((..............(((((((......)).))))).((((((.(.....).)))))).)))..))).)))...))))...)))....- ( -29.60) >DroWil_CAF1 93269 119 - 1 CUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCACGACAUGUGGGCCACCCAUAAAACUGAUAUCGGAGUAGUGCCAC- ......(((....((((..(((.(((...(((..............(((((((......)).))))).((((((((.....)))))))).)))..))).)))...))))...)))....- ( -33.60) >DroMoj_CAF1 83129 120 - 1 CUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACAUGUGGGCCACCCAUAAAACUGAUAUCGGAGUAGUGCCCCG ......(((....((((..(((.(((...(((..............(((((((......)).))))).((((((.(.....).)))))).)))..))).)))...))))...)))..... ( -29.60) >consensus CUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACAUGUGGGCCACCCAUAAAACUGAUAUCGGAGUAGUGCCUC_ ......(((....((((..(((.(((...(((..............(((((((......)).))))).((((((.(.....).)))))).)))..))).)))...))))...)))..... (-27.88 = -28.38 + 0.50)


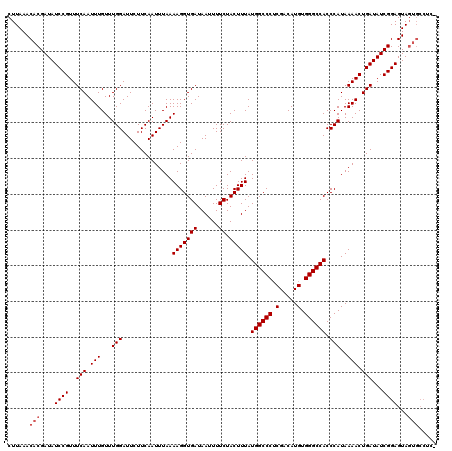
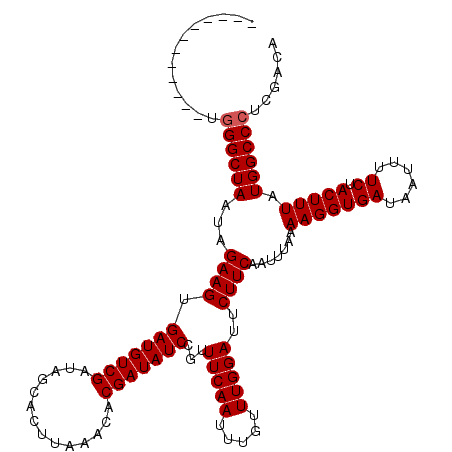
| Location | 3,594,406 – 3,594,514 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.36 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -22.81 |
| Energy contribution | -22.97 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3594406 108 - 22407834 ------------AGGGCUAAUAGAAGUGAUGUCGAUAGCACUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACA ------------(((((((...((((.(((((((..............)))))))...(((((.....)))))..)))).......(((((((......)).))))).)))))))..... ( -24.24) >DroVir_CAF1 74895 108 - 1 ------------UGGGCUAAUAGAAGUGAUGUCGAUAGCACUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACA ------------.((((((...((((.(((((((..............)))))))...(((((.....)))))..)))).......(((((((......)).))))).))))))...... ( -23.74) >DroGri_CAF1 76551 108 - 1 ------------UGGGCUAAUAGAAGUGAUGUCGAUAGCACUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACA ------------.((((((...((((.(((((((..............)))))))...(((((.....)))))..)))).......(((((((......)).))))).))))))...... ( -23.74) >DroWil_CAF1 93308 120 - 1 UUCUGUUUCCUUUUGGCUAAUAGAAGUGAUGUCGAUAGCACUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCACGACA (((((((.((....))..)))))))....(((((...............((.(((((...(.....)..)))))...)).........(((.((((.........)))).)))..))))) ( -22.20) >DroMoj_CAF1 83169 108 - 1 ------------UGGGCUAAUAGAAGUGAUGUCGAUAGCACUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACA ------------.((((((...((((.(((((((..............)))))))...(((((.....)))))..)))).......(((((((......)).))))).))))))...... ( -23.74) >DroPer_CAF1 69066 108 - 1 ------------AGGGCUAAUAGAAGUGAUGUCGAUAGCACUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACA ------------(((((((...((((.(((((((..............)))))))...(((((.....)))))..)))).......(((((((......)).))))).)))))))..... ( -24.24) >consensus ____________UGGGCUAAUAGAAGUGAUGUCGAUAGCACUUAAACACGAUAUCCGUUUCAAUUUGUUUGGAUUCUUCAAUUUAAAAGGUGAUAAUUUUCUACUUUAUGGCCCUCGACA .............((((((...((((.(((((((..............)))))))...(((((.....)))))..)))).......(((((((......)).))))).))))))...... (-22.81 = -22.97 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:31 2006