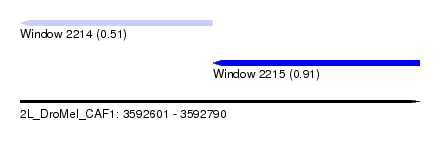
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,592,601 – 3,592,790 |
| Length | 189 |
| Max. P | 0.913397 |

| Location | 3,592,601 – 3,592,692 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -17.52 |
| Consensus MFE | -12.90 |
| Energy contribution | -12.77 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3592601 91 - 22407834 ------AA-GGCAACACGA--------GGAAGAGAAACACUUGAGUGGCACAUAAAAGUCACAUCGACAACU--UCGAUUGACUAAAGAAAAUAUUGUACAUG-------AACAA----- ------..-.((.((.(((--------(...........)))).)).)).......(((((.(((((.....--))))))))))..........((((.....-------.))))----- ( -15.80) >DroPse_CAF1 66530 107 - 1 G--AGAGA-GGCAACAUGG--------GGAAGAGAAACACUUGAGUGGCAUAUAAAAGUCACAUCGACAACU--UCGAUUGACUAAAGAAAAUAUUGUACAUGGCGAUGGAACACUGAAC .--.....-(....)....--------.............((.((((.........(((((.(((((.....--))))))))))........((((((.....))))))...)))).)). ( -18.40) >DroEre_CAF1 62157 91 - 1 ------AA-GGCAACACGA--------GGAAGGGAAACACUUGAGUGGCACAUAAAAGUCACAUCGACAACU--UCGAUUGACUAAAGAAAAUAUUGUACAUG-------AACAA----- ------..-.((.((.(((--------(....(....).)))).)).)).......(((((.(((((.....--))))))))))..........((((.....-------.))))----- ( -18.70) >DroWil_CAF1 90381 101 - 1 CCAACAUG-GACAACAUCGGCAACAUUGGGGCAGAAACACUUGAGUGGCAUAUAAAAGUCACAUCGACAACUUUUCGAUUGACUAAAGAAAAUAUUGUGUGU------------------ ((((.(((-.....))).(....).))))........(((....)))((((((((.(((((.(((((.......))))))))))..........))))))))------------------ ( -18.40) >DroAna_CAF1 65202 92 - 1 ------AAGAGCAACAUGA--------GGAAAAGAAACACUUGAGUGGCAUAUAAAAGUCACAUCGACAACU--UCGAUUGACUAAAGAAAAUAUUGUACAUG-------AACAA----- ------........((((.--------..........(((....))).........(((((.(((((.....--))))))))))...............))))-------.....----- ( -15.40) >DroPer_CAF1 66784 107 - 1 G--AGAGA-GGCAACAUGG--------GGAAGAGAAACACUUGAGUGGCAUAUAAAAGUCACAUCGACAACU--UCGAUUGACUAAAGAAAAUAUUGUACAUGGCGAUGGAACACUGAAC .--.....-(....)....--------.............((.((((.........(((((.(((((.....--))))))))))........((((((.....))))))...)))).)). ( -18.40) >consensus ______AA_GGCAACAUGA________GGAAGAGAAACACUUGAGUGGCAUAUAAAAGUCACAUCGACAACU__UCGAUUGACUAAAGAAAAUAUUGUACAUG_______AACAA_____ ..........((((.......................(((....))).........(((((.(((((.......))))))))))..........))))...................... (-12.90 = -12.77 + -0.14)

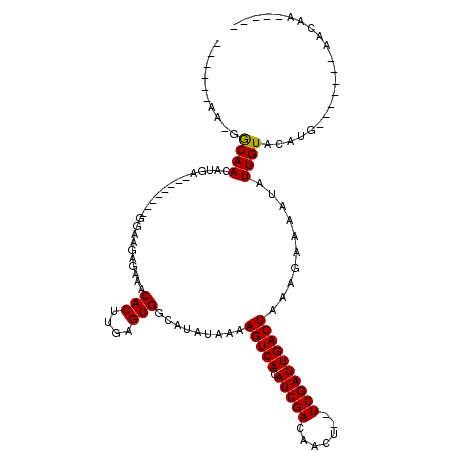
| Location | 3,592,692 – 3,592,790 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -19.02 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3592692 98 - 22407834 UCUCCAACAAUGAUAUUAAA--AAUAACAAUUGUCUUCAGUGGUCUUUAACACCCGCACCGCUGAUAGACAACCAAA---AACAGCAGAG-CAGCA-AAGCAGCA------ ....................--........(((((((((((((((..........).)))))))).)))))).....---....((...(-(....-..)).)).------ ( -18.70) >DroSim_CAF1 63738 89 - 1 UCUCCAACAAUGAUAUUAAA--AAUAACAAUUGUCUUCAGUGGUCUUUAACACCCGCACCGCUGAUAGACAAC-AAA---AACAGCAGAG-CA---------GCA------ ....................--........(((((((((((((((..........).)))))))).)))))).-...---....((....-..---------)).------ ( -18.00) >DroEre_CAF1 62248 96 - 1 UCUCCAACAAUGAUAUUAAA--AAUAACAAUUGUCUUCAGUGGUCUUUAACACCCGCACCGCUGAUAGGCAAC-A----AGGCAGCAAGG-CAGCA-AGGCAGCA------ .((((...............--........(((((((((((((((..........).)))))))).)))))).-.----..((.((...)-).)).-.)).))..------ ( -19.50) >DroWil_CAF1 90482 104 - 1 GCCUCAACAAUGAUAUUAAAAAAAUAACAAUUGUCUUCAAUGGUCUUUAACACCCGCACCGCUGAUAGACAAC-AGC---AACAACAUGGCCAACA-UGG--CCAACAUGG ((........((.((((.....)))).)).(((((((((.(((((..........).)))).))).)))))).-.))---.......(((((....-.))--)))...... ( -20.50) >DroYak_CAF1 63534 101 - 1 UCUCCAACAAUGAUAUUAAA--AAUAACAAUUGUCUUCAGUGGUCUUUAACACCCGCACCGCUGAUAGACAAC-AAAAAAAGCAGCAGAG-CAGCAAAGGCAGCA------ .((((...............--........(((((((((((((((..........).)))))))).)))))).-.......((.((...)-).))...)).))..------ ( -20.30) >DroAna_CAF1 65294 82 - 1 UCGCCAACAAUGAUAUUAAA--AAUAACAAUUGUCUUCAGUGGUCUUUAACACCCGCACCGCUGAUAGACAAC-AAAA--AACA------------------GCA------ ..((......((.(((....--.))).)).(((((((((((((((..........).)))))))).)))))).-....--....------------------)).------ ( -17.10) >consensus UCUCCAACAAUGAUAUUAAA__AAUAACAAUUGUCUUCAGUGGUCUUUAACACCCGCACCGCUGAUAGACAAC_AAA___AACAGCAGAG_CAGCA_AGG__GCA______ ..............................(((((((((((((((..........).)))))))).))))))....................................... (-15.70 = -15.73 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:28 2006