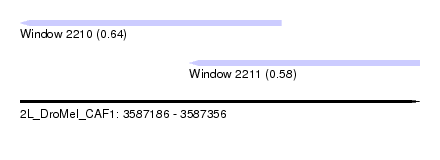
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,587,186 – 3,587,356 |
| Length | 170 |
| Max. P | 0.642295 |

| Location | 3,587,186 – 3,587,297 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
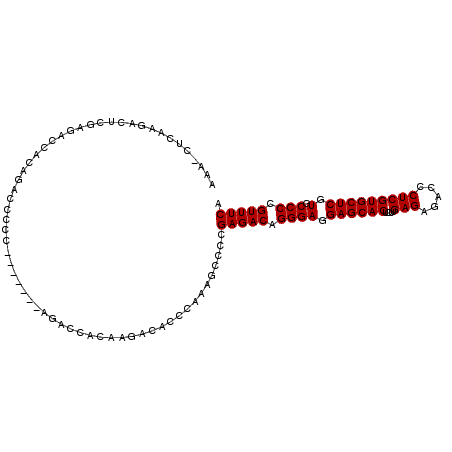
>2L_DroMel_CAF1 3587186 111 - 22407834 AAA-CUCAAGACUCAAGACCACAGACCCCCAGACCCCAGACCACAAGACACCCAAAGCCCCGAGACAGGGAGGAGCACUCUUGAGAGACCCUCGUGCUCGUGCCCCGUUUCA ...-.........................................................(((((.((((.((((((....(((.....))))))))).).))).))))). ( -23.30) >DroSec_CAF1 54913 104 - 1 AAA-CUCAAGACUCGAGACCACAUACCCCC-------AGACCACAAGACACCCAAAGCCCCGAGACAGGGAGGAGCACUCUUGAGAGACCCUCGUGCUCGUGCCCCGUUUCA ...-(((.......))).............-------........................(((((.((((.((((((....(((.....))))))))).).))).))))). ( -25.00) >DroSim_CAF1 58328 104 - 1 AAA-CUCAAGACUCGAGACCACAGAUCCCC-------AGACCACAAGACACCCAAAGCCCCGAGACAGGGAGGAGCACUCUUGAGAGACCCUCGUGCUCGUGCCCCGUUUCA ...-(((.......))).............-------........................(((((.((((.((((((....(((.....))))))))).).))).))))). ( -25.00) >DroAna_CAF1 59570 91 - 1 AAAACUAAAGA-----AAAAAC---------------UGAAAACU-GAAAACCCAGGCCUCGAGACAGGGAGGAGCACUCUUGAGAGACCCUCGUGCUCGUGCCCUGUUUCA ...........-----......---------------.((...((-(......)))...))((((((((((.((((((....(((.....))))))))).).))))))))). ( -30.70) >consensus AAA_CUCAAGACUCGAGACCACAGACCCCC_______AGACCACAAGACACCCAAAGCCCCGAGACAGGGAGGAGCACUCUUGAGAGACCCUCGUGCUCGUGCCCCGUUUCA .............................................................(((((.((((.((((((....(((.....))))))))).).))).))))). (-23.30 = -23.30 + 0.00)

| Location | 3,587,258 – 3,587,356 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 90.94 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -18.45 |
| Energy contribution | -19.23 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
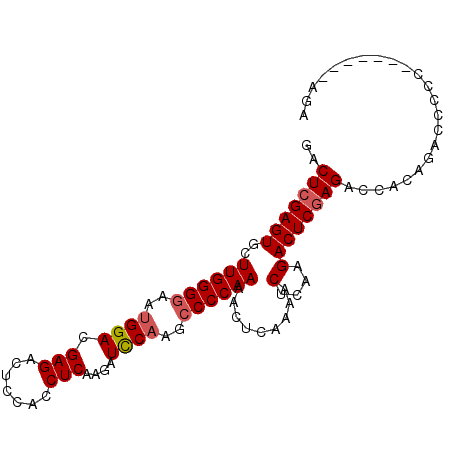
>2L_DroMel_CAF1 3587258 98 - 22407834 GACUCGAGUGCUUGGGGAAUGGACGAGACUCCACCUCAAGAUUCAAGCCCCAAAACUCAAAACUCAAGACUCAAGACCACAGACCCCCAGACCCCAGA .....((((..((((((..((((.(((.......)))....))))..)))))).))))........................................ ( -19.80) >DroSec_CAF1 54985 91 - 1 GACUCGAGUGCUUGGGAAAUGGACGAGACUCCACCUCAAGAUCCAAGCCCCAAAACUCAAAACUCAAGACUCGAGACCACAUACCCCC-------AGA ..(((((((.((((((...((((.(((.......)))....))))((........)).....))))))))))))).............-------... ( -21.50) >DroSim_CAF1 58400 91 - 1 GACUCGAGUGCUUGGGGAAUGGACGAGACUCCACCUCAAGAUCCCAGCCCCAAAACUCAAAACUCAAGACUCGAGACCACAGAUCCCC-------AGA ..(((((((..((((((...(((.(((.......)))....)))...)))))).........(....)))))))).............-------... ( -23.10) >consensus GACUCGAGUGCUUGGGGAAUGGACGAGACUCCACCUCAAGAUCCAAGCCCCAAAACUCAAAACUCAAGACUCGAGACCACAGACCCCC_______AGA ..(((((((..((((((..((((.(((.......)))....))))..)))))).........(....))))))))....................... (-18.45 = -19.23 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:24 2006