
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,579,679 – 3,579,799 |
| Length | 120 |
| Max. P | 0.728865 |

| Location | 3,579,679 – 3,579,799 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -46.32 |
| Consensus MFE | -33.63 |
| Energy contribution | -33.58 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
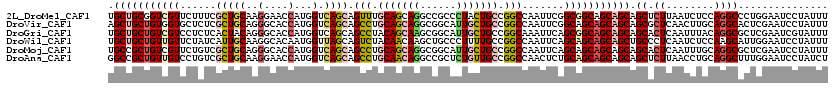
>2L_DroMel_CAF1 3579679 120 + 22407834 AAAUAGGAUUCCAGGGCCUGGAGAUUAAGAGCUGCUGCUGCCGCCGAAUUGGCCGGCAGUAGGGCGGCCUGCUGCAAACUGCUGACCAUGGUUCCUUGCAGCGAAAGAACGACCGCAGCA ........((((((...)))))).......(((((((((((((((.....)).))))))))((.((..(((((((((...(((......)))...)))))))...))..)).))))))). ( -48.40) >DroVir_CAF1 54731 120 + 1 AAAUAGGAUUCGAGUGCCUGCAAGUUGAGCGCUGCUGCUGCUGCCGAAUUGGCCGGCAGCAAUGCCGCCUGCUGCAGGCUGCUGACCAUGGUGCCCUGCAGCGAGAGCACCACAGCAGCU ......(.(((((.((....))..))))))(((((((.((((((((.......)))))))).....((((((((((((..(((......)))..)))))))).)).))....))))))). ( -52.90) >DroGri_CAF1 56059 120 + 1 AAAUACGAUUCGAGCGCCUGUAAAUUGAGUGCUGCUGCUGCCGCUGAAUUUGCCGGCAGCAAUGCCGCUUGCUGUAGGCUGCUGACCAUGGUGCCCUGUAGUGAGAGGACGACAGCAGCA ............((((((........).)))))(((((((.((((........(((((....)))))((..(((((((..(((......)))..)))))))..)).)).)).))))))). ( -45.40) >DroWil_CAF1 74291 120 + 1 AAAUAGGAUUCCAAUGCUUGGAGAUUGAGGGCAGCUGCUGCUGCUGAAUUGGCCGGCAAAAGGGCAGCUUGUUGUAGACUGCUAACCAUUGUGCCUUGCAAUGAUAGAACAACAGCAGCA ........(((((.....)))))..........(((((((.(((((.......)))))....(((((((......)).)))))...((((((.....)))))).........))))))). ( -34.30) >DroMoj_CAF1 62400 120 + 1 AAAUAGGAUUCGAGCGCCUGCAAAUUGAGUGCUGCUGCUGCUGCUGAAUUGGCCGGCAGCAAUGCCGCCUGCUGCAGGCUGCUGACCAUGGUGCCCUGCAGCGACAGAACGACAGCGGCA ............((((((........).)))))((((((((..(((....(((.((((....)))))))(((((((((..(((......)))..))))))))).)))...).))))))). ( -52.90) >DroAna_CAF1 51512 120 + 1 AGAUAGGAUUCCAAAGCCUGCAGGUUAAGAGCUGCUGCUGCUGCAGAGUUGGCCGGCAACAGAGCGGCCUGUUGCAGGCUGCUGACCAUGGUUCCUUGCAGCGACAGGACAACAGCGGCC ..............((((....))))....(((((((.((((((((....(((((.(......))))))..))))))(((((.(((....)))....))))).......)).))))))). ( -44.00) >consensus AAAUAGGAUUCCAGCGCCUGCAAAUUGAGAGCUGCUGCUGCUGCUGAAUUGGCCGGCAGCAAUGCCGCCUGCUGCAGGCUGCUGACCAUGGUGCCCUGCAGCGACAGAACGACAGCAGCA ...((((.........))))..........(((((((((((((((.....)).)))))))).........((((((((.((((......)))).))))))))............))))). (-33.63 = -33.58 + -0.05)


| Location | 3,579,679 – 3,579,799 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -45.60 |
| Consensus MFE | -30.78 |
| Energy contribution | -31.73 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3579679 120 - 22407834 UGCUGCGGUCGUUCUUUCGCUGCAAGGAACCAUGGUCAGCAGUUUGCAGCAGGCCGCCCUACUGCCGGCCAAUUCGGCGGCAGCAGCAGCUCUUAAUCUCCAGGCCCUGGAAUCCUAUUU .((((((((((......)(((((((.(.((....))...)...))))))).))))......((((((.((.....))))))))..)))))........(((((...)))))......... ( -42.70) >DroVir_CAF1 54731 120 - 1 AGCUGCUGUGGUGCUCUCGCUGCAGGGCACCAUGGUCAGCAGCCUGCAGCAGGCGGCAUUGCUGCCGGCCAAUUCGGCAGCAGCAGCAGCGCUCAACUUGCAGGCACUCGAAUCCUAUUU .((((((((((((((((......)))))))))))).)))).((((((((..((((((.((((((((((.....))))))))))..))..))))....))))))))............... ( -61.40) >DroGri_CAF1 56059 120 - 1 UGCUGCUGUCGUCCUCUCACUACAGGGCACCAUGGUCAGCAGCCUACAGCAAGCGGCAUUGCUGCCGGCAAAUUCAGCGGCAGCAGCAGCACUCAAUUUACAGGCGCUCGAAUCGUAUUU (((((((((((((((........))))).............(((..((((((......))))))..))).........))))))))))((.((........))))............... ( -40.00) >DroWil_CAF1 74291 120 - 1 UGCUGCUGUUGUUCUAUCAUUGCAAGGCACAAUGGUUAGCAGUCUACAACAAGCUGCCCUUUUGCCGGCCAAUUCAGCAGCAGCAGCUGCCCUCAAUCUCCAAGCAUUGGAAUCCUAUUU .((((((((((((.....((((...((((.((.((...((((.((......)))))))))).))))...))))..))))))))))))...........((((.....))))......... ( -36.50) >DroMoj_CAF1 62400 120 - 1 UGCCGCUGUCGUUCUGUCGCUGCAGGGCACCAUGGUCAGCAGCCUGCAGCAGGCGGCAUUGCUGCCGGCCAAUUCAGCAGCAGCAGCAGCACUCAAUUUGCAGGCGCUCGAAUCCUAUUU .((((((((.((((((..((((((((((((....))..))..)))))))).(((((((....)))).)))....))).))).))))).(((.......))).)))............... ( -48.40) >DroAna_CAF1 51512 120 - 1 GGCCGCUGUUGUCCUGUCGCUGCAAGGAACCAUGGUCAGCAGCCUGCAACAGGCCGCUCUGUUGCCGGCCAACUCUGCAGCAGCAGCAGCUCUUAACCUGCAGGCUUUGGAAUCCUAUCU ((((((((((((...((.(((((..(....)...).)))).(((.((((((((....)))))))).)))..))...)))))))).((((........)))).)))).............. ( -44.60) >consensus UGCUGCUGUCGUUCUCUCGCUGCAAGGCACCAUGGUCAGCAGCCUGCAGCAGGCGGCACUGCUGCCGGCCAAUUCAGCAGCAGCAGCAGCACUCAAUCUGCAGGCACUCGAAUCCUAUUU .(((((((((((......(((((..(....)...).)))).(((.(((((((......))))))).))).......))))))))))).((.((........))))............... (-30.78 = -31.73 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:18 2006