
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,579,263 – 3,579,400 |
| Length | 137 |
| Max. P | 0.999758 |

| Location | 3,579,263 – 3,579,374 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.15 |
| Mean single sequence MFE | -53.62 |
| Consensus MFE | -32.21 |
| Energy contribution | -32.52 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.60 |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3579263 111 + 22407834 CG---CACUCUGCUGCCAGUUCUGCGGCUGUUGCGCAGUCCAGCGCCGCCUGUUGGCUCGCAGCAGCUGCUGCGGCAGCCGCAGCAGCCGCCACUGCAGCACUAACGGUGCCAA .(---((((.((((((.(((...(((((((((((((......))((((((....)))..(((((....))))))))....))))))))))).))))))))).....)))))... ( -61.90) >DroPse_CAF1 51511 111 + 1 CG---CACUCGGCGGCCAGCUCUGCGGCUGUUGCACAGUCGAGGGCCGCUUGUUGAUUGGCCGCGGCCGCAGCGGCGGCAGCAGCUGCUGCCACAGCAGCACUCACAGUUCCAA .(---(.....((((((((.((.((((((.(((......))).)))))).....)))))))))).(((((....))))).)).(((((((...))))))).............. ( -54.50) >DroSec_CAF1 47013 111 + 1 CG---CACUCUGCUGCGAGUUCUGCGGCUGUUGCGCAGUCCAGCGCUGCCUGUUGGCUCGCAGCAGCUGCUGCGGCAGCCGCAGCAGCCGCCACUGCAGCACUAACGGUGCCAA .(---((((.((((((.(((...((((((((((((((((.....))))).....((((((((((....))))))..))))))))))))))).))))))))).....)))))... ( -63.00) >DroWil_CAF1 73896 90 + 1 CGGCACAUUCUGCAGCCAAUUCGGCGG------CACAAUCUAAUGCUGCCUGUUGAUUGGCAGCAACUGCAGCAGCUG------------------CCGCACUCACUGUACCGA (((((....((((.(((((((.(((((------((........)))))))....))))))).((....)).)))).))------------------)))............... ( -34.20) >DroPer_CAF1 51618 111 + 1 CG---CACUCGGCGGCCAGCUCUGCGGCUGUUGCACAGUCGAGGGCCGCUUGUUGAUUGGCCGCGGCCGCAGCGGCGGCAGCAGCUGCUGCCACAGCAGCACUCACAGUUCCAA .(---(.....((((((((.((.((((((.(((......))).)))))).....)))))))))).(((((....))))).)).(((((((...))))))).............. ( -54.50) >consensus CG___CACUCUGCGGCCAGUUCUGCGGCUGUUGCACAGUCCAGCGCCGCCUGUUGAUUGGCAGCAGCUGCAGCGGCAGCAGCAGCAGCCGCCACAGCAGCACUCACAGUGCCAA ...........((((((((.((.(((((....((((......)))).))).)).)))))))))).(((((....))))).((.((((......)))).)).............. (-32.21 = -32.52 + 0.31)


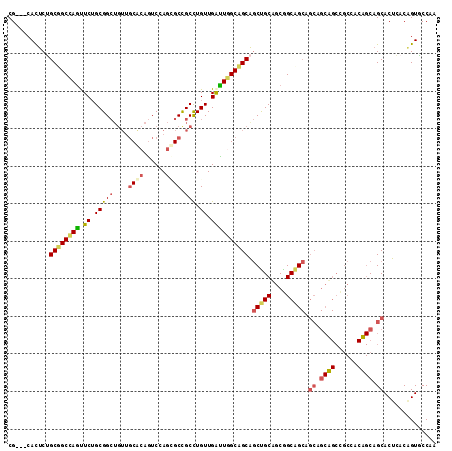
| Location | 3,579,263 – 3,579,374 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.15 |
| Mean single sequence MFE | -53.20 |
| Consensus MFE | -35.33 |
| Energy contribution | -37.52 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3579263 111 - 22407834 UUGGCACCGUUAGUGCUGCAGUGGCGGCUGCUGCGGCUGCCGCAGCAGCUGCUGCGAGCCAACAGGCGGCGCUGGACUGCGCAACAGCCGCAGAACUGGCAGCAGAGUG---CG ...(((((((((((.((((.(..(((((((((((((...)))))))))))))..).........(((.((((......))))....))))))).))))))....).)))---). ( -62.60) >DroPse_CAF1 51511 111 - 1 UUGGAACUGUGAGUGCUGCUGUGGCAGCAGCUGCUGCCGCCGCUGCGGCCGCGGCCAAUCAACAAGCGGCCCUCGACUGUGCAACAGCCGCAGAGCUGGCCGCCGAGUG---CG ............(..((((.((((((((....)))))))).)).((((((((((((...........)))).....((((((....).))))).)).))))))..))..---). ( -50.60) >DroSec_CAF1 47013 111 - 1 UUGGCACCGUUAGUGCUGCAGUGGCGGCUGCUGCGGCUGCCGCAGCAGCUGCUGCGAGCCAACAGGCAGCGCUGGACUGCGCAACAGCCGCAGAACUCGCAGCAGAGUG---CG ...(((((.....((((((.(..(((((((((((((...)))))))))))))..)(((......(((.((((......))))....)))......)))))))))).)))---). ( -60.80) >DroWil_CAF1 73896 90 - 1 UCGGUACAGUGAGUGCGG------------------CAGCUGCUGCAGUUGCUGCCAAUCAACAGGCAGCAUUAGAUUGUG------CCGCCGAAUUGGCUGCAGAAUGUGCCG .((((((((..(.(((((------------------(....)))))).)..))((((((.....(((.((((......)))------).)))..))))))........)))))) ( -41.40) >DroPer_CAF1 51618 111 - 1 UUGGAACUGUGAGUGCUGCUGUGGCAGCAGCUGCUGCCGCCGCUGCGGCCGCGGCCAAUCAACAAGCGGCCCUCGACUGUGCAACAGCCGCAGAGCUGGCCGCCGAGUG---CG ............(..((((.((((((((....)))))))).)).((((((((((((...........)))).....((((((....).))))).)).))))))..))..---). ( -50.60) >consensus UUGGAACCGUGAGUGCUGCAGUGGCAGCAGCUGCGGCCGCCGCUGCAGCUGCUGCCAAUCAACAGGCGGCCCUCGACUGUGCAACAGCCGCAGAACUGGCAGCAGAGUG___CG .............((((((.(((((.((....)).))))).)))))).((((((((((......(((.((((......))))....)))......))))))))))......... (-35.33 = -37.52 + 2.19)


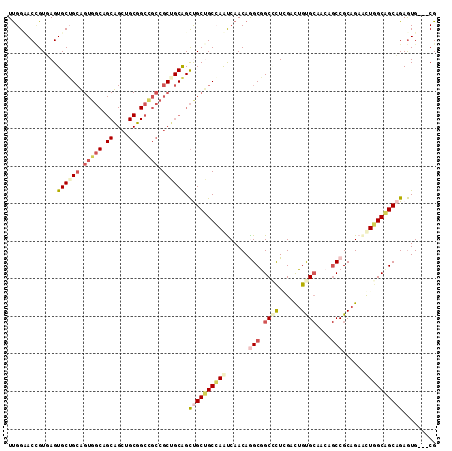
| Location | 3,579,300 – 3,579,400 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 67.22 |
| Mean single sequence MFE | -43.08 |
| Consensus MFE | -22.91 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3579300 100 + 22407834 CAGCGCCGCCUGUUGGCUCGCAGCAGCUGCUGCGGCAGCCGCAGCAGCCGCCACUGCAGCACUAACGGUGCCAAUGGAGGGCAGAGGCACGCACUUGGAU----------- ..(((..((((....(((((((((.((((((((((...)))))))))).))...))).((((.....)))).......))))..)))).)))........----------- ( -46.80) >DroPse_CAF1 51548 111 + 1 GAGGGCCGCUUGUUGAUUGGCCGCGGCCGCAGCGGCGGCAGCAGCUGCUGCCACAGCAGCACUCACAGUUCCAAUCGAAGGCAGUGGCAGGCACUUGGUGGCACAUGAAGU (...((((((..((((((((..((.(((((....))))).)).(((((((...)))))))..........))))))))....))))))...)((((.(((...))).)))) ( -47.60) >DroSec_CAF1 47050 100 + 1 CAGCGCUGCCUGUUGGCUCGCAGCAGCUGCUGCGGCAGCCGCAGCAGCCGCCACUGCAGCACUAACGGUGCCAAUGGAGGGCAGAGGCAGGCACUUGGAU----------- (((.(.((((((((.(((((((((.((((((((((...)))))))))).))...))).((((.....)))).......))))...))))))))))))...----------- ( -50.10) >DroWil_CAF1 73930 82 + 1 UAAUGCUGCCUGUUGAUUGGCAGCAACUGCAGCAGCUG------------------CCGCACUCACUGUACCGAUUGAGGGUAAUUUUGUUCGAUUUGUU----------- ...(((((((........)))))))..(((.((....)------------------).)))........((.(((((((.(......).))))))).)).----------- ( -23.30) >DroPer_CAF1 51655 111 + 1 GAGGGCCGCUUGUUGAUUGGCCGCGGCCGCAGCGGCGGCAGCAGCUGCUGCCACAGCAGCACUCACAGUUCCAAUCGAAGGCAGUGGCAGGCACUUGGUGGCACAUGAAGU (...((((((..((((((((..((.(((((....))))).)).(((((((...)))))))..........))))))))....))))))...)((((.(((...))).)))) ( -47.60) >consensus CAGCGCCGCCUGUUGAUUGGCAGCAGCUGCAGCGGCAGCAGCAGCAGCCGCCACAGCAGCACUCACAGUGCCAAUCGAGGGCAGUGGCAGGCACUUGGUU___________ ....(((((((.((((((((..((.(((((....))))).)).((((......)))).............))))))))))))...)))....................... (-22.91 = -23.62 + 0.71)

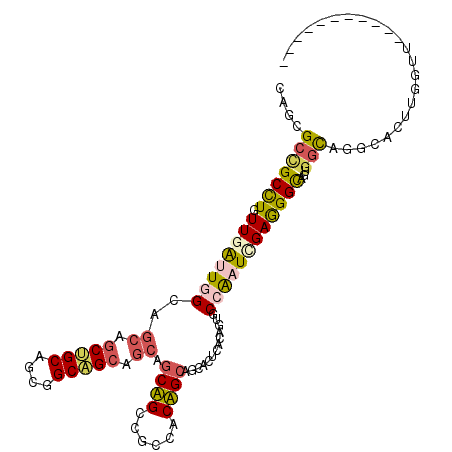
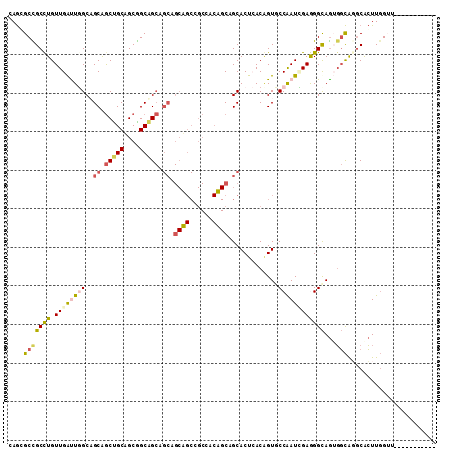
| Location | 3,579,300 – 3,579,400 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 67.22 |
| Mean single sequence MFE | -44.32 |
| Consensus MFE | -25.49 |
| Energy contribution | -26.72 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3579300 100 - 22407834 -----------AUCCAAGUGCGUGCCUCUGCCCUCCAUUGGCACCGUUAGUGCUGCAGUGGCGGCUGCUGCGGCUGCCGCAGCAGCUGCUGCGAGCCAACAGGCGGCGCUG -----------.....((((((((((..((.....))..)))))(((..((...((.(..(((((((((((((...)))))))))))))..)..))..))..)))))))). ( -54.30) >DroPse_CAF1 51548 111 - 1 ACUUCAUGUGCCACCAAGUGCCUGCCACUGCCUUCGAUUGGAACUGUGAGUGCUGCUGUGGCAGCAGCUGCUGCCGCCGCUGCGGCCGCGGCCAAUCAACAAGCGGCCCUC .........(((....((((.....))))((....((((((..(((((..(((.((.((((((((....)))))))).)).)))..))))))))))).....))))).... ( -44.20) >DroSec_CAF1 47050 100 - 1 -----------AUCCAAGUGCCUGCCUCUGCCCUCCAUUGGCACCGUUAGUGCUGCAGUGGCGGCUGCUGCGGCUGCCGCAGCAGCUGCUGCGAGCCAACAGGCAGCGCUG -----------.....(((((.(((((.....(((....(((((.....)))))...(..(((((((((((((...)))))))))))))..)))).....)))))))))). ( -54.30) >DroWil_CAF1 73930 82 - 1 -----------AACAAAUCGAACAAAAUUACCCUCAAUCGGUACAGUGAGUGCGG------------------CAGCUGCUGCAGUUGCUGCCAAUCAACAGGCAGCAUUA -----------.................((((.......))))....((.(((((------------------(....)))))).))((((((........)))))).... ( -24.60) >DroPer_CAF1 51655 111 - 1 ACUUCAUGUGCCACCAAGUGCCUGCCACUGCCUUCGAUUGGAACUGUGAGUGCUGCUGUGGCAGCAGCUGCUGCCGCCGCUGCGGCCGCGGCCAAUCAACAAGCGGCCCUC .........(((....((((.....))))((....((((((..(((((..(((.((.((((((((....)))))))).)).)))..))))))))))).....))))).... ( -44.20) >consensus ___________AACCAAGUGCCUGCCACUGCCCUCCAUUGGAACCGUGAGUGCUGCAGUGGCAGCAGCUGCGGCCGCCGCUGCAGCUGCUGCCAAUCAACAGGCGGCCCUC ............................((((.......))))....(((((((((.(((((.((....)).))))).(((((....)))))..........))))))))) (-25.49 = -26.72 + 1.23)

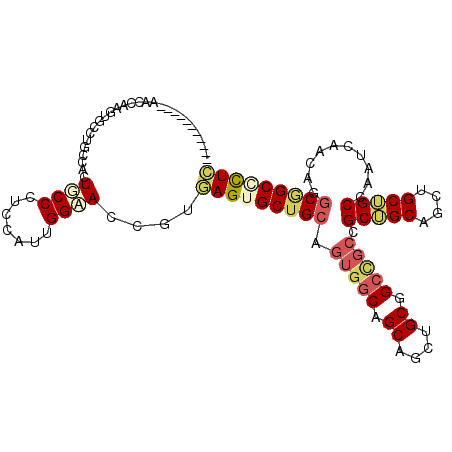

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:16 2006