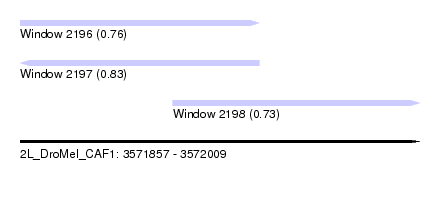
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,571,857 – 3,572,009 |
| Length | 152 |
| Max. P | 0.831917 |

| Location | 3,571,857 – 3,571,948 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -22.83 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3571857 91 + 22407834 UGGAGAUGGGCACUUUAAAGUGCCCCGU-AAACUGUCAACGGCAGCCCGAAGGGAACCGAAAACGGUUGUCGGGGUUACCGG------UACCAGACUU ..(((..(((((((....)))))))...-...(((....(((.(((((...((.(((((....))))).)).))))).))).------...))).))) ( -35.70) >DroSec_CAF1 39754 91 + 1 UCGAGAUGGGCACUUUAAAGUGCCCCGU-GAACUGUCAACGGCAGCCCGAAGGGAUCCGAAAACGGUUGUCGGGGUUACCGG------UACCGGACUU (((.((((((((((....)))))))...-...((((.....)))).((....))))))))...(((.(..(((.....))).------.))))..... ( -33.10) >DroSim_CAF1 40618 91 + 1 UCAAGAUGGGCACUUUAAAGUGCCCCGU-GAACUGUCAACGGCAGCCCGAAGGGAUCCGAAAACGGUUGUCGGGGUUACCGG------UACCGGACUU (((....(((((((....)))))))..)-))...(((..(((...(((...)))..)))....(((.(..(((.....))).------.))))))).. ( -33.80) >DroEre_CAF1 41312 83 + 1 UCGAGAUGGGCACUUUAAAGUGCCCCGU-GAACUGUCAACGGCAGCCCGAAGGGAUCCGAAAACGGUGGUCGGGAU--------------CCGGAUUC (((.((((((((((....)))))))...-...((((.....))))(((((......(((....)))...)))))))--------------)))).... ( -29.50) >DroYak_CAF1 41923 97 + 1 UCGAGAUGGGCACUUUAAAGUGCCCCGU-GAACUGUCAACGGCAGCCCGAGGGGAUCCGAAAACGGUUGUCGGGGUUACCGGUACUGGUACCGGAAUU (((....(((((((....)))))))..)-)).((((.....))))(((((.((...(((....))))).)))))....((((((....)))))).... ( -37.60) >DroAna_CAF1 44234 73 + 1 ------UGAGCACUUUAAACUGCCCCGCAGAACUGUCAACGGCAGCCCGAAAGGGA-------------UCGGGGUUAUGGA------UUCCGGAUUU ------.............(((((..(((....)))....)))))(((....))).-------------((((((((...))------)))))).... ( -22.10) >consensus UCGAGAUGGGCACUUUAAAGUGCCCCGU_GAACUGUCAACGGCAGCCCGAAGGGAUCCGAAAACGGUUGUCGGGGUUACCGG______UACCGGACUU .......(((((((....))))))).......((((.....))))(((((......(((....)))...)))))........................ (-22.83 = -23.28 + 0.46)



| Location | 3,571,857 – 3,571,948 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -21.85 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3571857 91 - 22407834 AAGUCUGGUA------CCGGUAACCCCGACAACCGUUUUCGGUUCCCUUCGGGCUGCCGUUGACAGUUU-ACGGGGCACUUUAAAGUGCCCAUCUCCA ....(((.((------.(((((..(((((.(((((....)))))....))))).))))).)).)))...-...(((((((....)))))))....... ( -34.10) >DroSec_CAF1 39754 91 - 1 AAGUCCGGUA------CCGGUAACCCCGACAACCGUUUUCGGAUCCCUUCGGGCUGCCGUUGACAGUUC-ACGGGGCACUUUAAAGUGCCCAUCUCGA ....(((...------.)))......(((.(((.(((..(((...((....))...)))..))).))).-...(((((((....)))))))...))). ( -31.30) >DroSim_CAF1 40618 91 - 1 AAGUCCGGUA------CCGGUAACCCCGACAACCGUUUUCGGAUCCCUUCGGGCUGCCGUUGACAGUUC-ACGGGGCACUUUAAAGUGCCCAUCUUGA ....(((...------.)))......((..(((.(((..(((...((....))...)))..))).))).-.))(((((((....)))))))....... ( -31.00) >DroEre_CAF1 41312 83 - 1 GAAUCCGG--------------AUCCCGACCACCGUUUUCGGAUCCCUUCGGGCUGCCGUUGACAGUUC-ACGGGGCACUUUAAAGUGCCCAUCUCGA .....(((--------------((..((...((.(((..(((...((....))...)))..))).))..-.))(((((((....)))))))))).)). ( -28.50) >DroYak_CAF1 41923 97 - 1 AAUUCCGGUACCAGUACCGGUAACCCCGACAACCGUUUUCGGAUCCCCUCGGGCUGCCGUUGACAGUUC-ACGGGGCACUUUAAAGUGCCCAUCUCGA ....((((((....))))))......(((.(((.(((..(((..(((...)))...)))..))).))).-...(((((((....)))))))...))). ( -34.80) >DroAna_CAF1 44234 73 - 1 AAAUCCGGAA------UCCAUAACCCCGA-------------UCCCUUUCGGGCUGCCGUUGACAGUUCUGCGGGGCAGUUUAAAGUGCUCA------ ....((((((------(...(((((((((-------------......))))).....))))...)))))).)(((((.(....).))))).------ ( -18.80) >consensus AAGUCCGGUA______CCGGUAACCCCGACAACCGUUUUCGGAUCCCUUCGGGCUGCCGUUGACAGUUC_ACGGGGCACUUUAAAGUGCCCAUCUCGA ....((((........))))....(((((...(((....)))......)))))....(((.((....)).)))(((((((....)))))))....... (-21.85 = -22.53 + 0.68)


| Location | 3,571,915 – 3,572,009 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.65 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -14.69 |
| Energy contribution | -16.42 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3571915 94 + 22407834 AAAACGGUUGUCGGGGUUACCGG------UACCAGACUUCGGAAUCCGGAAUCCGGGU---UAUA---------UAUUUGGUUAUAUGCUUCGUUGAUCUUUGA-------ACGCAAAA .....(((((.((((((......------.(((((((....)(((((((...))))))---)...---------..)))))).....)))))).))))).....-------........ ( -23.30) >DroSec_CAF1 39812 94 + 1 AAAACGGUUGUCGGGGUUACCGG------UACCGGACUUCGGAAUCCGGAAUCCGGGU---UAUA---------UAUUUAGUUAUAUGCUUCGUUGAUCUUUGA-------ACGCAAAA .....(((((.((((((..((((------..((((((....)..)))))...))))..---.(((---------((......))))))))))).))))).....-------........ ( -25.90) >DroSim_CAF1 40676 94 + 1 AAAACGGUUGUCGGGGUUACCGG------UACCGGACUUCGGAAUCCGGAAUCCGGGU---UAUA---------UAUUUGGUUAUAUGCUUCGUUGAUCUUUAA-------ACGCAAAA .....(((((.((((((..((((------..((((((....)..)))))...))))..---.(((---------((......))))))))))).))))).....-------........ ( -25.90) >DroEre_CAF1 41370 86 + 1 AAAACGGUGGUCGGGAU--------------CCGGAUUCCGGAUUCCGGAAUCCGGGA---CAUA---------UAUUCGGUUAUAUGCUUCGUUGAUCUUUGA-------ACGCAAAA ......(((.(((((((--------------(((((((((((...))))))))))(((---((((---------((......)))))).)))...)))).))))-------.))).... ( -32.10) >DroYak_CAF1 41981 109 + 1 AAAACGGUUGUCGGGGUUACCGGUACUGGUACCGGAAUUCAGAGUCCGGAAUCCGGGA---UAUACAUACAUAUUAUUUGGUUAUAUGCUUCGUUGAUCUUUGA-------ACGCAAAA .....(((((.((((((.(((((..((((..(((((........)))))...))))((---(((......)))))..))))).....)))))).))))).....-------........ ( -25.80) >DroAna_CAF1 44284 93 + 1 ----------UCGGGGUUAUGGA------UUCCGGAUUUCGGAUUCCGUAUUCCGGGUGGAUAUA---------UAUUU-UUUAUAUGCAUCGUUGAUCUUUCAAUGGUUAACAAAAAA ----------.((((((.(((((------.((((.....)))).)))))))))))((((.(((((---------.....-..))))).))))(((((((.......)))))))...... ( -33.10) >consensus AAAACGGUUGUCGGGGUUACCGG______UACCGGACUUCGGAAUCCGGAAUCCGGGU___UAUA_________UAUUUGGUUAUAUGCUUCGUUGAUCUUUGA_______ACGCAAAA .....(((((.((((((..............(((((.(((((...))))).)))))..................(((......))).)))))).))))).................... (-14.69 = -16.42 + 1.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:12 2006