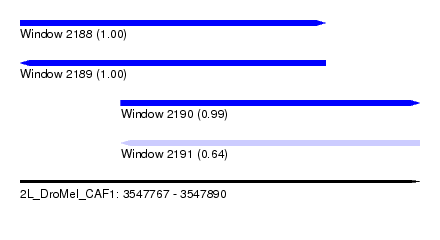
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,547,767 – 3,547,890 |
| Length | 123 |
| Max. P | 0.999863 |

| Location | 3,547,767 – 3,547,861 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -20.80 |
| Energy contribution | -20.68 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.30 |
| SVM RNA-class probability | 0.999863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3547767 94 + 22407834 AUGCAGCAUACAGCAUGCGACGCGGAUGCAAAAUGCAGCAUGCAACGUGCGACAACUAUCAGCUGCAAUUAUCUUGCCUUAAAACAUAUUACCG .((((((.....((((.((...)).))))....((..(((((...)))))..)).......))))))........................... ( -22.90) >DroSec_CAF1 16704 94 + 1 AUGCAGCAUGCAACAUGCGACACGCCAGCAAAAUGCAGCAUGCAACAUGCGACAACUAUCAGCUGCAAUUAUCUUGCCUUAAAACAUAUUACCG .((((((.((((...(((.........)))...))))(((((...)))))...........))))))........................... ( -22.00) >DroSim_CAF1 17001 94 + 1 AUGCAGCAUGCAACAUGCGACACGCCAGCAACAUGCAGCAUGCAACAUGCGACAACUAUCAGCUGCAAUUAUCUUGCCUUAAAACAUAUUACCG .((((((.((((...(((.........)))...))))(((((...)))))...........))))))........................... ( -22.00) >DroEre_CAF1 16922 94 + 1 AUGCGGCAUGCAACAUGCGUCACACCAGCAACAUGCAGCAUGCAACAUGCGACAACUAUCAGCUGCAAAUAUCUUGCCUUAAAACAUAUUACCG .((((((..((.....))(((......((.....)).(((((...))))))))........))))))........................... ( -22.00) >DroYak_CAF1 17410 94 + 1 AUGCAGCAUGCAACAUGCGACACACCAGCAACAUGCAGCAUGCAACAUGCGACAACUAUCAGCUGCAAUUAUCUUGCCUUAAAACAUAUUACCG .((((((.((((...(((.........)))...))))(((((...)))))...........))))))........................... ( -22.00) >consensus AUGCAGCAUGCAACAUGCGACACGCCAGCAACAUGCAGCAUGCAACAUGCGACAACUAUCAGCUGCAAUUAUCUUGCCUUAAAACAUAUUACCG .((((((.((((...(((.........)))...))))(((((...)))))...........))))))........................... (-20.80 = -20.68 + -0.12)

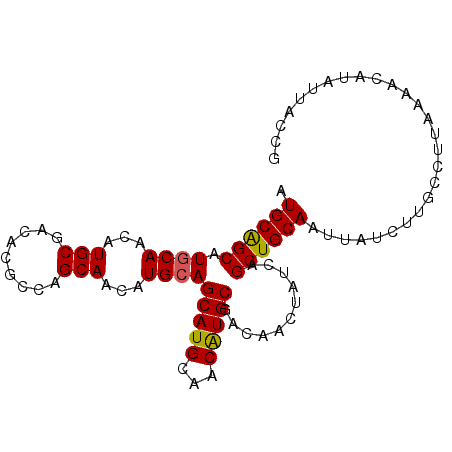
| Location | 3,547,767 – 3,547,861 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -27.10 |
| Energy contribution | -27.58 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3547767 94 - 22407834 CGGUAAUAUGUUUUAAGGCAAGAUAAUUGCAGCUGAUAGUUGUCGCACGUUGCAUGCUGCAUUUUGCAUCCGCGUCGCAUGCUGUAUGCUGCAU ........((((....)))).......((((((..(((((((.((.((((.(.((((........))))).)))))))).)))))..)))))). ( -26.30) >DroSec_CAF1 16704 94 - 1 CGGUAAUAUGUUUUAAGGCAAGAUAAUUGCAGCUGAUAGUUGUCGCAUGUUGCAUGCUGCAUUUUGCUGGCGUGUCGCAUGUUGCAUGCUGCAU ........((((....)))).......((((((.......(((.((((((.(((((((((.....)).))))))).)))))).))).)))))). ( -31.10) >DroSim_CAF1 17001 94 - 1 CGGUAAUAUGUUUUAAGGCAAGAUAAUUGCAGCUGAUAGUUGUCGCAUGUUGCAUGCUGCAUGUUGCUGGCGUGUCGCAUGUUGCAUGCUGCAU .................(((.(((((((((....).))))))))((((((.((((((.((((((.....)))))).)))))).))))))))).. ( -34.50) >DroEre_CAF1 16922 94 - 1 CGGUAAUAUGUUUUAAGGCAAGAUAUUUGCAGCUGAUAGUUGUCGCAUGUUGCAUGCUGCAUGUUGCUGGUGUGACGCAUGUUGCAUGCCGCAU (((((((((((......((((.((....(((((.....)))))...)).)))).....)))))))))))(((((..(((((...)))))))))) ( -30.40) >DroYak_CAF1 17410 94 - 1 CGGUAAUAUGUUUUAAGGCAAGAUAAUUGCAGCUGAUAGUUGUCGCAUGUUGCAUGCUGCAUGUUGCUGGUGUGUCGCAUGUUGCAUGCUGCAU .................(((.(((((((((....).))))))))((((((.((((((.(((..(.....)..))).)))))).))))))))).. ( -31.30) >consensus CGGUAAUAUGUUUUAAGGCAAGAUAAUUGCAGCUGAUAGUUGUCGCAUGUUGCAUGCUGCAUGUUGCUGGCGUGUCGCAUGUUGCAUGCUGCAU ((..(((.((((.....((((.....))))....)))))))..)).((((.((((((.((((((.((......)).)))))).)))))).)))) (-27.10 = -27.58 + 0.48)

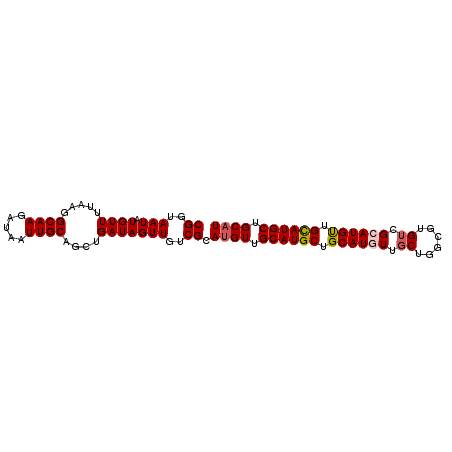
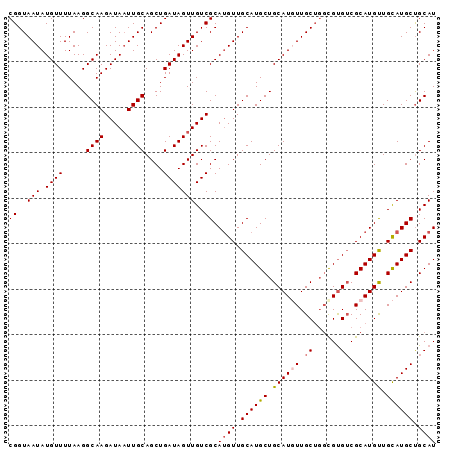
| Location | 3,547,798 – 3,547,890 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -11.40 |
| Consensus MFE | -11.40 |
| Energy contribution | -11.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3547798 92 + 22407834 AAUGCAGCAUGCAACGUGCGACAACUAUCAGCUGCAAUUAUCUUGCCUUAAAACAUAUUACCGAUUUACACACAUACACACACACUCUCGCU- ..((((((..((.....))((......)).))))))........................................................- ( -11.40) >DroSec_CAF1 16735 91 + 1 AAUGCAGCAUGCAACAUGCGACAACUAUCAGCUGCAAUUAUCUUGCCUUAAAACAUAUUACCGAUUUACACACAUAAAC--ACACUCUUGCUA ..((((((..((.....))((......)).))))))...........................................--............ ( -11.40) >DroSim_CAF1 17032 91 + 1 CAUGCAGCAUGCAACAUGCGACAACUAUCAGCUGCAAUUAUCUUGCCUUAAAACAUAUUACCGAUUUACAUACAUACAC--ACACUCUCGCUA ..((((((..((.....))((......)).))))))...........................................--............ ( -11.40) >DroEre_CAF1 16953 91 + 1 CAUGCAGCAUGCAACAUGCGACAACUAUCAGCUGCAAAUAUCUUGCCUUAAAACAUAUUACCGACUUACACACAUACAC--ACACUCUCGCUA ..((((((..((.....))((......)).))))))...........................................--............ ( -11.40) >DroYak_CAF1 17441 91 + 1 CAUGCAGCAUGCAACAUGCGACAACUAUCAGCUGCAAUUAUCUUGCCUUAAAACAUAUUACCGACUUACACACAUACAC--ACACUAUCGCUA ..((((((..((.....))((......)).))))))...........................................--............ ( -11.40) >consensus CAUGCAGCAUGCAACAUGCGACAACUAUCAGCUGCAAUUAUCUUGCCUUAAAACAUAUUACCGAUUUACACACAUACAC__ACACUCUCGCUA ..((((((..((.....))((......)).))))))......................................................... (-11.40 = -11.40 + 0.00)

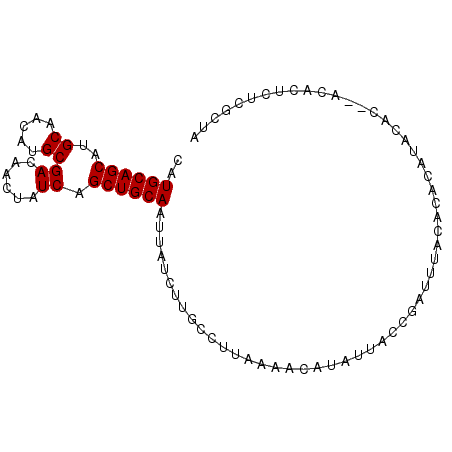
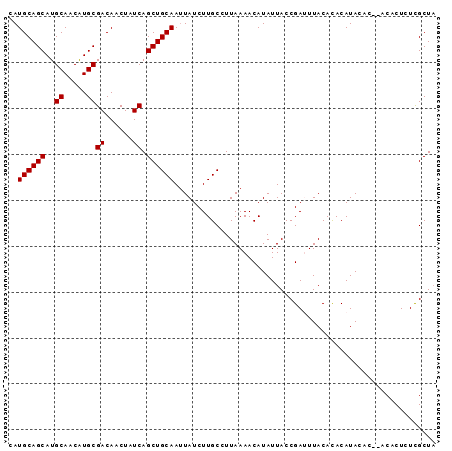
| Location | 3,547,798 – 3,547,890 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -22.06 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.54 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3547798 92 - 22407834 -AGCGAGAGUGUGUGUGUAUGUGUGUAAAUCGGUAAUAUGUUUUAAGGCAAGAUAAUUGCAGCUGAUAGUUGUCGCACGUUGCAUGCUGCAUU -......(((((((((((((((((((((((((((..((.....))..((((.....)))).))))))..))).)))))).))))))).))))) ( -23.70) >DroSec_CAF1 16735 91 - 1 UAGCAAGAGUGU--GUUUAUGUGUGUAAAUCGGUAAUAUGUUUUAAGGCAAGAUAAUUGCAGCUGAUAGUUGUCGCAUGUUGCAUGCUGCAUU ..(((...((((--((..((((((((((((((((..((.....))..((((.....)))).))))))..))).))))))).)))))))))... ( -20.90) >DroSim_CAF1 17032 91 - 1 UAGCGAGAGUGU--GUGUAUGUAUGUAAAUCGGUAAUAUGUUUUAAGGCAAGAUAAUUGCAGCUGAUAGUUGUCGCAUGUUGCAUGCUGCAUG ..((....))..--(((((.((((((((..((..(((.((((.....((((.....))))....)))))))..))....))))))))))))). ( -23.30) >DroEre_CAF1 16953 91 - 1 UAGCGAGAGUGU--GUGUAUGUGUGUAAGUCGGUAAUAUGUUUUAAGGCAAGAUAUUUGCAGCUGAUAGUUGUCGCAUGUUGCAUGCUGCAUG ..((...(((((--(((.((((((((((((((((...(((((((.....))))))).....))))))..))).)))))))))))))))))... ( -21.20) >DroYak_CAF1 17441 91 - 1 UAGCGAUAGUGU--GUGUAUGUGUGUAAGUCGGUAAUAUGUUUUAAGGCAAGAUAAUUGCAGCUGAUAGUUGUCGCAUGUUGCAUGCUGCAUG ....(.((((((--(((.((((((((((((((((..((.....))..((((.....)))).))))))..))).)))))))))))))))))... ( -21.20) >consensus UAGCGAGAGUGU__GUGUAUGUGUGUAAAUCGGUAAUAUGUUUUAAGGCAAGAUAAUUGCAGCUGAUAGUUGUCGCAUGUUGCAUGCUGCAUG ..((....))....(((((.((((((((..((..(((.((((.....((((.....))))....)))))))..))....))))))))))))). (-19.50 = -19.54 + 0.04)

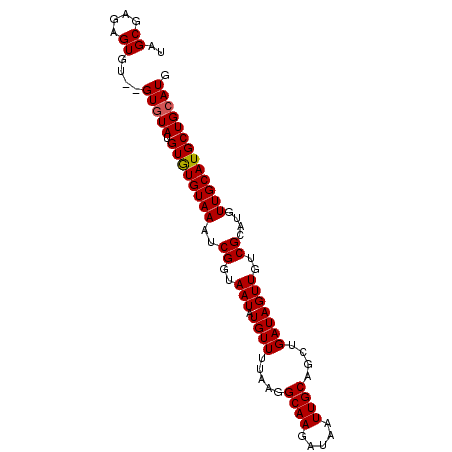
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:06 2006