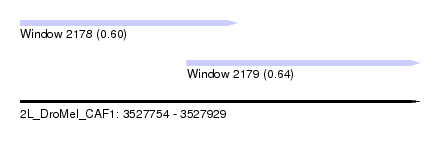
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,527,754 – 3,527,929 |
| Length | 175 |
| Max. P | 0.636455 |

| Location | 3,527,754 – 3,527,849 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.32 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -19.83 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3527754 95 + 22407834 GAAUCUUCGGCGAAGACUUGCCGUUUUUUGCUGAGGAUAGAGCGGAUAUUUUCGUUGGUCGAGUCUUUGAUGACGGGAAUCCUCACAGGAGUUCC ..((((((((((((((........))))))))))))))..((((((....))))))......(((......)))((((.(((.....))).)))) ( -30.10) >DroSec_CAF1 36765 95 + 1 GAGUCUUCGGCGAAGACUUGCCGUUUUUUGCUGAGGAUGGAGCGGAUAUUUUCGUUGGUCGAGUCUUUGAUUACGGAAAUCCUCACAGGAGUUCC ..((((((((((((((........))))))))))))))(((((.....(((((((.((((((....)))))))))))))(((.....)))))))) ( -32.90) >DroSim_CAF1 38434 95 + 1 GAGUCUUCGGCAAAGACUUGCCGUUUUUUGCUGAGGAUGGAGCGGAUAUUUUCGUUGGUCGAGUCCUUGAUUACGGAAAUCCUCACAGGAGUUCC ..((((((((((((((........))))))))))))))(((((.....(((((((.((((((....)))))))))))))(((.....)))))))) ( -34.60) >DroEre_CAF1 37897 92 + 1 GCAUCUUCGGCGAAGACUUGCCGUUUUUUGCUGAGGAUGGAACGCAAGUUUUCGU---UCGAGUUUUUGCUGACGGGAAUUCUCACAGGAGUUCC ....(.((((((((((((((((((((((....))))))))(((....))).....---.)))))))))))))).)((((((((....)))))))) ( -34.70) >DroYak_CAF1 36343 95 + 1 GAAUCUUCGCCGAAGACUUGGCGUUUUUUGCUGAGGAUGGAACGCACAUUAGCGUUGGUCGAGUCUUUGAUGAUGGGAAUCCUCACAGGAGUUCC ......(((.((((((((((((((((((....))))))..(((((......))))).)))))))))))).))).((((.(((.....))).)))) ( -31.80) >DroAna_CAF1 33064 95 + 1 GAGUCUUCCGCGAAAAGUUCCCUCUUUUUGCAUAGAAUCGAUCGUAGAUUGUUAUUAGUUGCGUCCUUUAUGACUGGAUUUCGGACAGGAGUGCC ..((.((((((((((((......))))))))...(((((.((((((((.(((........)))...))))))).).)))))......)))).)). ( -22.40) >consensus GAAUCUUCGGCGAAGACUUGCCGUUUUUUGCUGAGGAUGGAGCGGAUAUUUUCGUUGGUCGAGUCUUUGAUGACGGGAAUCCUCACAGGAGUUCC ..((((((((((((((........))))))))))))))..((((((....))))))......(((......)))((((.(((.....))).)))) (-19.83 = -20.72 + 0.89)

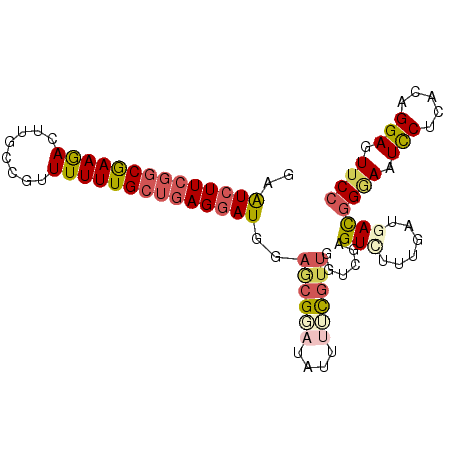
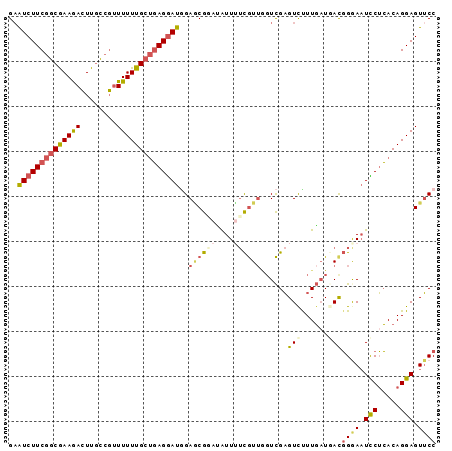
| Location | 3,527,827 – 3,527,929 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -25.53 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3527827 102 + 22407834 CGGGAAUCCUCACAGGAGUUCCUUUGCGCUGCGGUUUUUGUGGCGUUGGACUGUCCAGAAUUUCCACACUCAAUGUCCCAUCUUGCUUGAUGGGAGAUUCCU .(((((((......(((((((....((((..((.....))..))))(((.....))))))))))...........(((((((......)))))))))))))) ( -36.30) >DroSec_CAF1 36838 102 + 1 CGGAAAUCCUCACAGGAGUUCCUUUACGCUGCGGUUUUUGUGGCGUUGGACUGUCCAGGAUUUCAACGCUCAAAGUCCCAUCUGGCUUGAUGGGAGAUUCCU ..((((((((.((((....(((...((((..((.....))..)))).)))))))..))))))))...........(((((((......)))))))....... ( -36.70) >DroSim_CAF1 38507 102 + 1 CGGAAAUCCUCACAGGAGUUCCUUUACGCUGCGGUUUUUGUGGCGUUGGACUGUCCAGGAUUUCAACGCUCAAAGUCCCAUCUGCCUUGAUGGGAGAUUCCU ..((((((((.((((....(((...((((..((.....))..)))).)))))))..))))))))...........(((((((......)))))))....... ( -36.70) >DroEre_CAF1 37967 102 + 1 CGGGAAUUCUCACAGGAGUUCCUUUGCGCUGAGGCUUUUGUGGCGAGGGACUGUCCAGCAUUUCAACGAUCAAAGUGCCAUCUGGCUUGAUGGGAGAUUCCU .((((((.(((...(((..((((((((((..........)).))))))))...)))..........(.(((((...(((....)))))))).)))))))))) ( -32.30) >DroYak_CAF1 36416 102 + 1 UGGGAAUCCUCACAGGAGUUCCUUUGCGCUGAGGUUUUUGUGGCGUGGAACUGUCCAGGAUUUCAACGCUCAGAGUUCCAUCUGGCUUGAUGGGAGAUUCUU .(((((((((((..((((((((...(((((...........))))))))))..)))......((((.((.((((......)))))))))))))).))))))) ( -33.10) >DroAna_CAF1 33137 95 + 1 CUGGAUUUCGGACAGGAGUGCCCUUGCGCUGCGGUUUCUGUGGCGUGGGGCUGUCCAGGAUUUGUAU-----AGCUUCCGCCUUGCCAGGUGUAACAUCC-- ((((.....((((((......(((..(((..(((...)))..)))..))))))))).(((...(...-----..).)))......))))...........-- ( -34.70) >consensus CGGGAAUCCUCACAGGAGUUCCUUUGCGCUGCGGUUUUUGUGGCGUGGGACUGUCCAGGAUUUCAACGCUCAAAGUCCCAUCUGGCUUGAUGGGAGAUUCCU ..((((((((....(((((((....(((((((((...))))))))).))))..)))))))))))...........(((((((......)))))))....... (-25.53 = -25.82 + 0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:54 2006