
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,523,717 – 3,523,865 |
| Length | 148 |
| Max. P | 0.908492 |

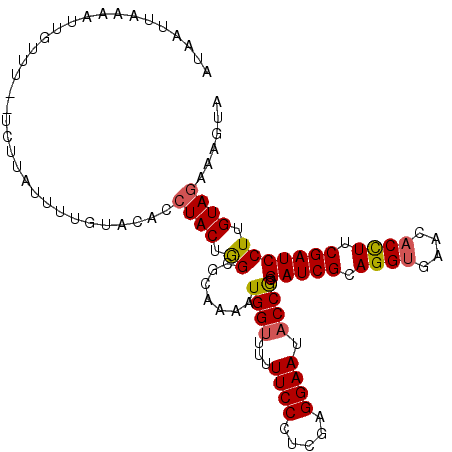
| Location | 3,523,717 – 3,523,825 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -19.68 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
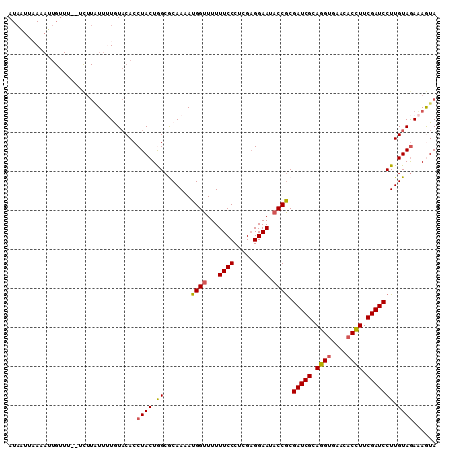
>2L_DroMel_CAF1 3523717 108 + 22407834 AUAAUUAAAAUUGUUU--UCUUAUUUUGUACACCUACUGGCGCAAAAUGGUUUUUUCCCUCGAGGAAUACCGCGAUCGCAGGUGAACACUUUCGAUCCUUGUAGAAAGUA .............(((--(((.(((((((...((....)).)))))))(((...((((.....)))).)))..(((((.((((....)))).))))).....)))))).. ( -22.40) >DroSec_CAF1 32728 108 + 1 AUAAUUUAAAUUGUUU--UCUUAUUUUGUACACCUACUGGCGCAAAAUGGUUUUUUCCCUCGAGGAAUACCGCGAUCGCAGGUGAACACCUUCGAUCCUUGUAGAAAGUA .............(((--(((.(((((((...((....)).)))))))(((...((((.....)))).)))..(((((.((((....)))).))))).....)))))).. ( -25.10) >DroSim_CAF1 34421 108 + 1 AUAAUUUAAAUUGUUU--UCUUAUUUUGUACACCUACUGGCGCAAAAUGGUUUUUUCCCUCGAGGAAUACCGCGAUCGCAGGCGAACACCUUCGAUCCUUGUAGAAAGUA .............(((--(((.(((((((...((....)).)))))))(((...((((.....)))).)))..(((((.(((......))).))))).....)))))).. ( -23.00) >DroEre_CAF1 30654 109 + 1 AUAGU-AAAAUUGUUUAUUUUUGUUAUAUACACCUACUAGCACAAAAUGGUUUUUUCCCUCGCGGAAUACCGCGAUCGCAGGUGAACACCUUCGAUCCUUGUAAAGAAUA .((((-(....(((.(((.......))).)))..)))))..........(((((((.....((((....))))(((((.((((....)))).))))).....))))))). ( -23.10) >DroYak_CAF1 32112 95 + 1 AUUAC-AAA--------------UAAUAUUCACCUACUAGCACAAAAUGGUUUUUUCCCUCGAGGAAUUCCACGAUCGCAGGUGAACACCUUCGAUCCUUGUAGAAAAUA .....-...--------------..........((((.((.......(((....((((.....))))..))).(((((.((((....)))).))))))).))))...... ( -21.00) >consensus AUAAUUAAAAUUGUUU__UCUUAUUUUGUACACCUACUGGCGCAAAAUGGUUUUUUCCCUCGAGGAAUACCGCGAUCGCAGGUGAACACCUUCGAUCCUUGUAGAAAGUA .................................((((.((.......((((...((((.....)))).)))).(((((.((((....)))).))))))).))))...... (-19.68 = -19.72 + 0.04)

| Location | 3,523,717 – 3,523,825 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -21.36 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3523717 108 - 22407834 UACUUUCUACAAGGAUCGAAAGUGUUCACCUGCGAUCGCGGUAUUCCUCGAGGGAAAAAACCAUUUUGCGCCAGUAGGUGUACAAAAUAAGA--AAACAAUUUUAAUUAU .((((((..........))))))((.(((((((...((((((.((((.....))))...))).....)))...))))))).)).........--................ ( -24.90) >DroSec_CAF1 32728 108 - 1 UACUUUCUACAAGGAUCGAAGGUGUUCACCUGCGAUCGCGGUAUUCCUCGAGGGAAAAAACCAUUUUGCGCCAGUAGGUGUACAAAAUAAGA--AAACAAUUUAAAUUAU ...(((((.....(((((.((((....)))).)))))..(((.((((.....))))...)))....((((((....)))))).......)))--)).............. ( -27.30) >DroSim_CAF1 34421 108 - 1 UACUUUCUACAAGGAUCGAAGGUGUUCGCCUGCGAUCGCGGUAUUCCUCGAGGGAAAAAACCAUUUUGCGCCAGUAGGUGUACAAAAUAAGA--AAACAAUUUAAAUUAU ...(((((.....(((((.((((....)))).)))))..(((.((((.....))))...)))....((((((....)))))).......)))--)).............. ( -27.60) >DroEre_CAF1 30654 109 - 1 UAUUCUUUACAAGGAUCGAAGGUGUUCACCUGCGAUCGCGGUAUUCCGCGAGGGAAAAAACCAUUUUGUGCUAGUAGGUGUAUAUAACAAAAAUAAACAAUUUU-ACUAU .......(((((((((((.((((....)))).)))))((((....))))..((.......))..)))))).((((((.(((.(((.......))).)))...))-)))). ( -27.60) >DroYak_CAF1 32112 95 - 1 UAUUUUCUACAAGGAUCGAAGGUGUUCACCUGCGAUCGUGGAAUUCCUCGAGGGAAAAAACCAUUUUGUGCUAGUAGGUGAAUAUUA--------------UUU-GUAAU .......((((((.......(((((((((((((..(((.((....)).)))((.......))...........))))))))))))).--------------)))-))).. ( -27.20) >consensus UACUUUCUACAAGGAUCGAAGGUGUUCACCUGCGAUCGCGGUAUUCCUCGAGGGAAAAAACCAUUUUGCGCCAGUAGGUGUACAAAAUAAGA__AAACAAUUUUAAUUAU .((((((..........))))))((.(((((((..(((.((....)).)))((.......))...........))))))).))........................... (-21.36 = -20.60 + -0.76)

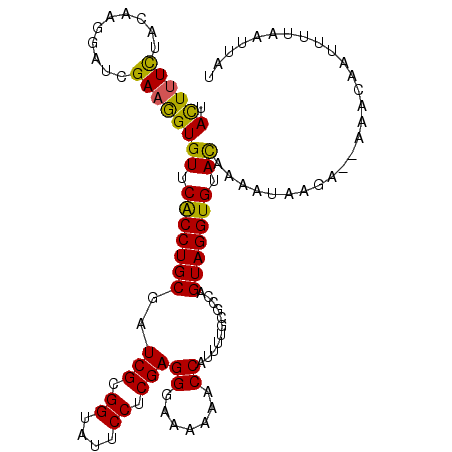
| Location | 3,523,745 – 3,523,865 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -27.92 |
| Energy contribution | -27.32 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
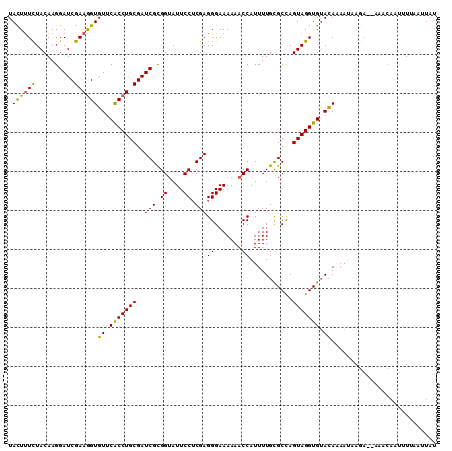
>2L_DroMel_CAF1 3523745 120 - 22407834 AAUAGUAAAUGGAUCCAUUAUUUUUCAUUGCGAUCAAGGAUACUUUCUACAAGGAUCGAAAGUGUUCACCUGCGAUCGCGGUAUUCCUCGAGGGAAAAAACCAUUUUGCGCCAGUAGGUG ....((((((((..............(((((((((.(((((((((((..........))))))))...)))..))))))))).((((.....))))....))).)))))(((....))). ( -34.10) >DroSec_CAF1 32756 120 - 1 AAUAGUUAAUGGAUCUAUUGUUUUCCAUUGCGAUCAAGGAUACUUUCUACAAGGAUCGAAGGUGUUCACCUGCGAUCGCGGUAUUCCUCGAGGGAAAAAACCAUUUUGCGCCAGUAGGUG .......((((((..........))))))((((((.(((((((((((..........))))))))...)))..))))))(((.((((.....))))...)))......((((....)))) ( -33.50) >DroSim_CAF1 34449 120 - 1 AAUAGUUAAUGGAUCUAUUGUUUUCCAUUGCGAUCAAGGAUACUUUCUACAAGGAUCGAAGGUGUUCGCCUGCGAUCGCGGUAUUCCUCGAGGGAAAAAACCAUUUUGCGCCAGUAGGUG .......((((((..........))))))((((((..((((((((((..........))))))))))(....)))))))(((.((((.....))))...)))......((((....)))) ( -34.10) >DroEre_CAF1 30683 120 - 1 AAAAUUAAAUGGAUCUAUACUUUUUCUUUGCGAUCAAGGAUAUUCUUUACAAGGAUCGAAGGUGUUCACCUGCGAUCGCGGUAUUCCGCGAGGGAAAAAACCAUUUUGUGCUAGUAGGUG ................((((((((((((((..(..((.....))..)..))))))..)))))))).(((((((..((((((....))))))((.......))...........))))))) ( -31.90) >DroYak_CAF1 32127 120 - 1 AAUAGUUAAUGGAUCCAUUGUUUUUCAUUGCGAUCAAGGAUAUUUUCUACAAGGAUCGAAGGUGUUCACCUGCGAUCGUGGAAUUCCUCGAGGGAAAAAACCAUUUUGUGCUAGUAGGUG ..((((.((((((((((((((........)))))...))))...((((((...(((((.((((....)))).)))))))))))((((.....))))....)))))....))))....... ( -31.90) >consensus AAUAGUUAAUGGAUCUAUUGUUUUUCAUUGCGAUCAAGGAUACUUUCUACAAGGAUCGAAGGUGUUCACCUGCGAUCGCGGUAUUCCUCGAGGGAAAAAACCAUUUUGCGCCAGUAGGUG .......(((((..............(((((((((.(((((((((((..........))))))))...)))..))))))))).((((....)))).....)))))...((((....)))) (-27.92 = -27.32 + -0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:52 2006