
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,479,846 – 3,479,937 |
| Length | 91 |
| Max. P | 0.730735 |

| Location | 3,479,846 – 3,479,937 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 85.58 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -24.08 |
| Energy contribution | -23.70 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
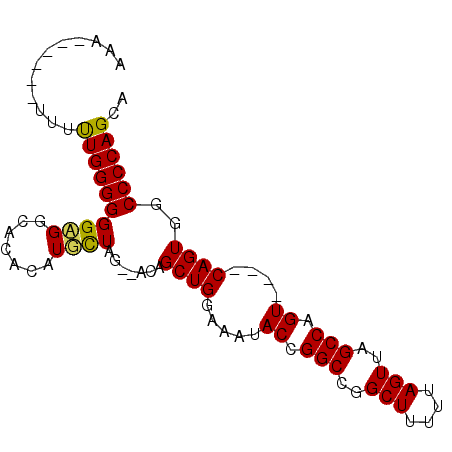
>2L_DroMel_CAF1 3479846 91 + 22407834 AAAGGGGGCUUUUUGGGGGGCGGUACACAUGUUAG--ACAGCUGGAAAUACCGGCCGGCUUUUUAGUUAGCCAGUCAGUCAGUGGCCCCAGCA ....((((((..((((..(((((((..((.(((..--..)))))....))))(((..(((....)))..))).)))..)))).)))))).... ( -33.40) >DroSim_CAF1 7019 89 + 1 AAAGGGGGCUUUUUGGGGGGCGGUACACAUGCUAGAGACAGCUGGAAAUACCGGCCGGCUUUUUAGUUAGCCAGU----CAGUGGCCCCAGCA ....((((((..((((.((.(((((..((.(((......)))))....))))).))((((........))))..)----))).)))))).... ( -35.00) >DroEre_CAF1 7336 81 + 1 AAA------UUUCUGGGGGGAGGCACGCAUUCUAG--ACAGCUGGAAAUACCGGCCGGCUUUUUAGUUAGCCAGU----CAGUGGCCCCAGCA ...------...(((((((((((....).)))).(--((.(((((.....))))).((((........)))).))----).....)))))).. ( -28.80) >DroYak_CAF1 7104 80 + 1 -CU------CUUUUGGGGGGAGGCACACAUGCUAG--GCAGCUGGAAAUACCGGCCGGCUUUUUAGUUAGCCAGU----CAGUGGCCCCAGCA -((------(....))).((.(((.(((..(((.(--((.(((((.....))))).((((....)))).))))))----..)))))))).... ( -28.40) >consensus AAA______UUUUUGGGGGGAGGCACACAUGCUAG__ACAGCUGGAAAUACCGGCCGGCUUUUUAGUUAGCCAGU____CAGUGGCCCCAGCA ............((((((((((.......)))).......((((.....((.(((..(((....)))..))).))....))))..)))))).. (-24.08 = -23.70 + -0.38)


| Location | 3,479,846 – 3,479,937 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 85.58 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -22.25 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3479846 91 - 22407834 UGCUGGGGCCACUGACUGACUGGCUAACUAAAAAGCCGGCCGGUAUUUCCAGCUGU--CUAACAUGUGUACCGCCCCCCAAAAAGCCCCCUUU .(((((((((((((...(((.((((........))))(((.((.....)).)))))--)...)).)))....)))))......)))....... ( -25.90) >DroSim_CAF1 7019 89 - 1 UGCUGGGGCCACUG----ACUGGCUAACUAAAAAGCCGGCCGGUAUUUCCAGCUGUCUCUAGCAUGUGUACCGCCCCCCAAAAAGCCCCCUUU ....(((((.....----...((((........))))(((.(((((...((((((....)))).)).)))))))).........))))).... ( -29.40) >DroEre_CAF1 7336 81 - 1 UGCUGGGGCCACUG----ACUGGCUAACUAAAAAGCCGGCCGGUAUUUCCAGCUGU--CUAGAAUGCGUGCCUCCCCCCAGAAA------UUU ..((((((..((((----.((((((........)))))).)))).......((...--.......))........))))))...------... ( -25.30) >DroYak_CAF1 7104 80 - 1 UGCUGGGGCCACUG----ACUGGCUAACUAAAAAGCCGGCCGGUAUUUCCAGCUGC--CUAGCAUGUGUGCCUCCCCCCAAAAG------AG- .(((((((..((((----.((((((........)))))).))))..)))))))...--...(((....))).............------..- ( -25.60) >consensus UGCUGGGGCCACUG____ACUGGCUAACUAAAAAGCCGGCCGGUAUUUCCAGCUGU__CUAGCAUGUGUACCGCCCCCCAAAAA______UUU ...(((((...........((((((........))))))..(((((...((((((....)))).)).)))))...)))))............. (-22.25 = -22.50 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:37 2006