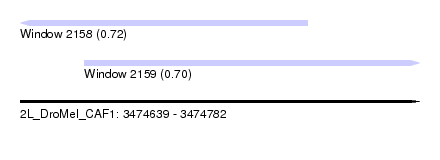
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,474,639 – 3,474,782 |
| Length | 143 |
| Max. P | 0.721571 |

| Location | 3,474,639 – 3,474,742 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.73 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -23.03 |
| Energy contribution | -24.58 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3474639 103 - 22407834 ACCAGAACUGAUUGCCACCCUGACUGUGGCAGCUCCACAUCCAAACGGUGGCAUUCGGCGGUCCCUCGUGCACAUCCAGGCACACUGAAUCGAAGCCCUUC------AU ..(((..((((.(((((((.((..(((((.....)))))..))...))))))).)))).........((((........)))).)))....(((....)))------.. ( -32.80) >DroSec_CAF1 1902 103 - 1 ACCAGAACUGAUUGCCACCCUGACUGUGGCAGUUCCACAUCCAGACGGUGGCAUUCGGCGGUCCCUCGUGCACAUCAAGGCAAACUGAGUCGAAGCCCUUC------AU ....(((..((.((((((((((..(((((.....)))))..)))..))))))).))(((..((.(((((((........)))...))))..)).))).)))------.. ( -34.60) >DroSim_CAF1 1923 103 - 1 ACCAGAACUGAUUGCCACCCUGACUGUGGCAGCUCCACAUCCAGACGGUGGCAUUCGGCGGUCCCUCGUGCACAUCCAGGCACACUGAGUCGAAGCCCUUC------AU ....(((..((.((((((((((..(((((.....)))))..)))..))))))).))(((..((.(((((((........))))...)))..)).))).)))------.. ( -36.40) >DroEre_CAF1 2311 103 - 1 ACCAGAACUGAUUGCCGCCCUGGGUGUGACAGUUCCACAUCCAAACGGUGGCAUUCGGCUGCGGCUCGUGGACAUCCAGGCACACUGAGUCCAAGCUCUUC------AU ..(((..((((.(((((((.((((((((.......))))))))...))))))).))))))).((((..(((((...(((.....))).)))))))))....------.. ( -41.70) >DroWil_CAF1 2589 100 - 1 ACCAAAAUUGAUUGCCACCUUGGUUAUGGCAAUUCCACAUCCAUAUGGUGGCAUUCGGAUGAUUAUCCUGAACAUCCAGGCAAUC---------GCCCUCCUCACCAAG .........((.(((((((.(((...(((.....)))...)))...))))))).))((.(((((..((((......)))).))))---------).))........... ( -31.10) >DroYak_CAF1 1907 103 - 1 ACCAGAACUGAUUGCCACCCUGACUGUGACAUUGCCACAUCCAAACGGUGGCAUCCGGCGGUCCCUCGUGCACAUCCAGGCACACUGAGUCGAAGCGCUUC------AU ..(((..(((..(((((((.((..((((.......))))..))...)))))))..))).........((((........)))).)))....(((....)))------.. ( -28.30) >consensus ACCAGAACUGAUUGCCACCCUGACUGUGGCAGUUCCACAUCCAAACGGUGGCAUUCGGCGGUCCCUCGUGCACAUCCAGGCACACUGAGUCGAAGCCCUUC______AU ..(((..((((.(((((((.((..(((((.....)))))..))...))))))).)))).........((((........)))).)))...................... (-23.03 = -24.58 + 1.56)

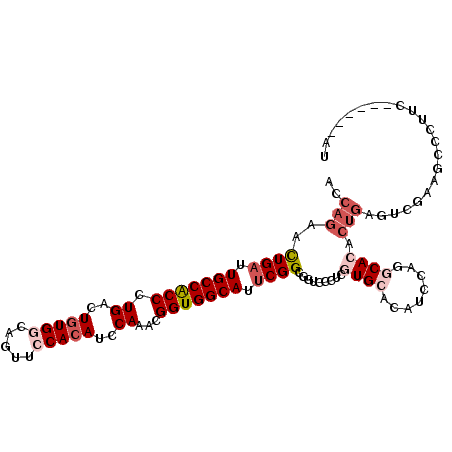
| Location | 3,474,662 – 3,474,782 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -41.82 |
| Consensus MFE | -27.99 |
| Energy contribution | -27.60 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3474662 120 + 22407834 CCUGGAUGUGCACGAGGGACCGCCGAAUGCCACCGUUUGGAUGUGGAGCUGCCACAGUCAGGGUGGCAAUCAGUUCUGGUACUAUGACAGGCAAACCCAAAGGUUGGUUCACGGAGAGAA ............((..((((((((((.(((((((..(((..(((((.....)))))..)))))))))).))(((......)))......))).((((....))))))))).))....... ( -41.00) >DroSec_CAF1 1925 120 + 1 CCUUGAUGUGCACGAGGGACCGCCGAAUGCCACCGUCUGGAUGUGGAACUGCCACAGUCAGGGUGGCAAUCAGUUCUGGUACUACGAUCGUCAAACCCAAAGGUUGGUGCACGGGGAGAA .(((..(((((((((.(((((((.((.(((((((..(((..(((((.....)))))..)))))))))).)).))...))).))....))....((((....)))).)))))))..))).. ( -47.20) >DroEre_CAF1 2334 120 + 1 CCUGGAUGUCCACGAGCCGCAGCCGAAUGCCACCGUUUGGAUGUGGAACUGUCACACCCAGGGCGGCAAUCAGUUCUGGUACUACGACCGGCAAACCCAACGAUUGGUUCACGGAGAAAC (((((.(((..(((..(((((.(((((((....))))))).)))))...))).))).))))).((..(((((((((((((......)))))..........))))))))..))....... ( -36.80) >DroWil_CAF1 2609 120 + 1 CCUGGAUGUUCAGGAUAAUCAUCCGAAUGCCACCAUAUGGAUGUGGAAUUGCCAUAACCAAGGUGGCAAUCAAUUUUGGUAUUACGAUCGAAAUCAUCAAUGGAUAGUUCACGGCAAGAA ((((((((((((((((....))))((.(((((((...(((.(((((.....))))).))).))))))).)).((((((((......))))))))......)))))).)))).))...... ( -37.60) >DroYak_CAF1 1930 120 + 1 CCUGGAUGUGCACGAGGGACCGCCGGAUGCCACCGUUUGGAUGUGGCAAUGUCACAGUCAGGGUGGCAAUCAGUUCUGGUACUACGACCGGUCAACCCAGAGGUUGAUUCACGGCGAGCA (((.(.......).)))(..((((((((((((((..(((..((((((...))))))..))))))))).)))....(((((......)))))((((((....))))))....)))))..). ( -46.60) >DroAna_CAF1 1909 120 + 1 CCUGGAUGUCCAGGACUCUAAUCCGAAUGCCACUGUCUGGAUGUGGCCGUGUCACGGCCAGGGCGGCAAUCAGUUCUGGUACUACGAUAGAAAGAAUCAGUGGCUGGUCCAUGGGAAAAA (((((....))))).(((....(((...((((((((((...((((((...))))))((((((((........))))))))............)))..)))))))))).....)))..... ( -41.70) >consensus CCUGGAUGUGCACGAGGGACAGCCGAAUGCCACCGUUUGGAUGUGGAACUGCCACAGCCAGGGUGGCAAUCAGUUCUGGUACUACGACCGGCAAACCCAAAGGUUGGUUCACGGAGAGAA (((((((.................((.(((((((..((((.(((((.....))))).))))))))))).))....(((((......)))))...............))))).))...... (-27.99 = -27.60 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:34 2006