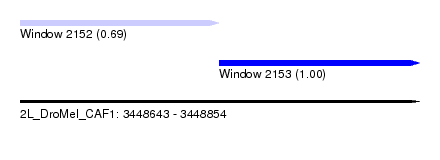
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,448,643 – 3,448,854 |
| Length | 211 |
| Max. P | 0.995626 |

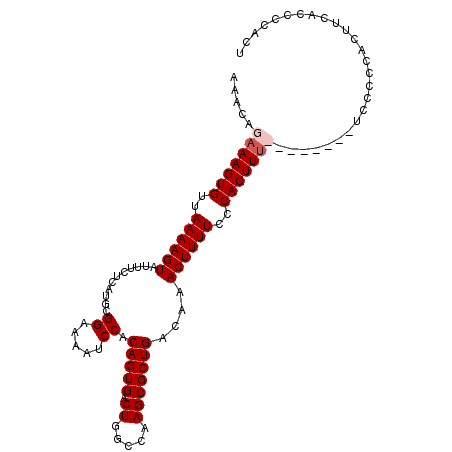
| Location | 3,448,643 – 3,448,748 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -25.48 |
| Energy contribution | -25.42 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3448643 105 + 22407834 GAGAUAAGGCCAGACAUCUGGGUGGGUUAUAAAUCUGGCGGGUUGGCCCGCUCAAAACCUGACCCUAUAAUCCUUUAUUUUCGUUUUCAAUUUGCUUGAAAAGGC (((((((((((((....))))..((((((.......((((((....)))))).......)))))).......)))))))))(.(((((((.....))))))).). ( -33.84) >DroSec_CAF1 15542 105 + 1 AAGAUAGGGCCAGACAUCUGGGUGGGUUAUAAAUCUGGCGGGCUGGCCCGCUCAAAACCUGAACCUAUAAUCCUUUAUUUUCAUUUUCAAUUUGUUGGAAAAGGC (((((((((((((....))))(((((((.......(((((((....))))).)).......)))))))....)))))))))..(((((((....))))))).... ( -31.94) >DroEre_CAF1 14789 105 + 1 GAGAUAAAGCCAGACAUCUGGAUGGGUAAUAAAUCUGGCGGUUCGGGUCGCUCAAAACCUGACCCUAUAAUCCUUUAUUUUCGUUUUCAAUUUGUUGGAAAAGGC ((((((((((((((.((((....))))......))))))((.((((((........)))))).))........))))))))..(((((((....))))))).... ( -24.80) >DroYak_CAF1 2740 105 + 1 AAGAUAAAGCCAGACAUCUGGGUGGGUUAUAAAUCUGGCGAUUUGGGCCGCUUAAAACCUGACCCUAUAAUCCUUUAUUUUCGUUUUCAAUUUGUUGGAAAAGGC (((((((((((((....))))..((((((.......((((........)))).......)))))).......)))))))))..(((((((....))))))).... ( -25.94) >consensus AAGAUAAAGCCAGACAUCUGGGUGGGUUAUAAAUCUGGCGGGUUGGCCCGCUCAAAACCUGACCCUAUAAUCCUUUAUUUUCGUUUUCAAUUUGUUGGAAAAGGC (((((((((((((....))))..((((((.......((((((....)))))).......)))))).......)))))))))..(((((((....))))))).... (-25.48 = -25.42 + -0.06)

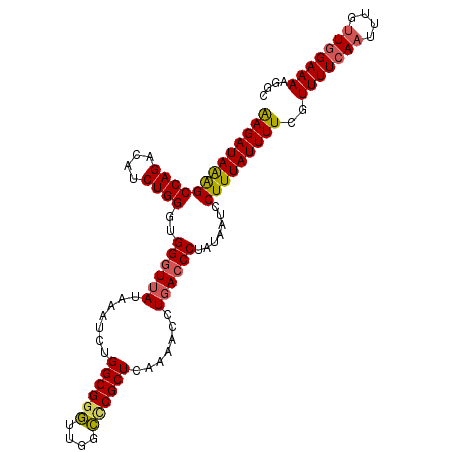
| Location | 3,448,748 – 3,448,854 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 89.97 |
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -19.98 |
| Energy contribution | -20.73 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3448748 106 + 22407834 AAACAGAAAGUGUUAAAAGUAUUUCUCAUGCGGAAAAUCCACAGUGACUGGCCAAGCGCUGACAAACUUUUCCCACUUUCCACACUUUUCCCCACUUCACCCCACU .....(((((((..((((((...........((.....)).(((((.((.....)))))))....))))))..))))))).......................... ( -22.90) >DroSec_CAF1 15647 106 + 1 AAACAGAAAGUGUUAAAAGUAUUUCUCAUGCGGAAAAUCCACAGUGACUGGCCAAGCGCUGACAAACUUUUCCCACUUUCCACACUUUUCCCCACUUCAGCCCACU .....(((((((..((((((...........((.....)).(((((.((.....)))))))....))))))..))))))).......................... ( -22.90) >DroEre_CAF1 14894 97 + 1 AAACAGAAAGUGUUAAAAGUAUUUCUCAUGCGGAAAAUCCACAGUGACUGGCCAAGCGCUGACAAACUUUUCCCACUU---------UCCCCCACUUUUCUCCGCU .....(((((((..((((((...........((.....)).(((((.((.....)))))))....))))))..)))))---------))................. ( -22.90) >DroYak_CAF1 2845 98 + 1 AAACAGAAAGUGUUAAAAGUAUUUCUCAUGCGGAAAAUCCACAGUGACUGGCCAAGCGCUGACAAACUUUUCCCACUUU--------UCCCCCACUUUUUCACCCU .....(((((((..((((((...........((.....)).(((((.((.....)))))))....))))))..))))))--------).................. ( -20.70) >consensus AAACAGAAAGUGUUAAAAGUAUUUCUCAUGCGGAAAAUCCACAGUGACUGGCCAAGCGCUGACAAACUUUUCCCACUUU________UCCCCCACUUCACCCCACU .....(((((((..((((((...........((.....)).(((((.((.....)))))))....))))))..))))))).......................... (-19.98 = -20.73 + 0.75)

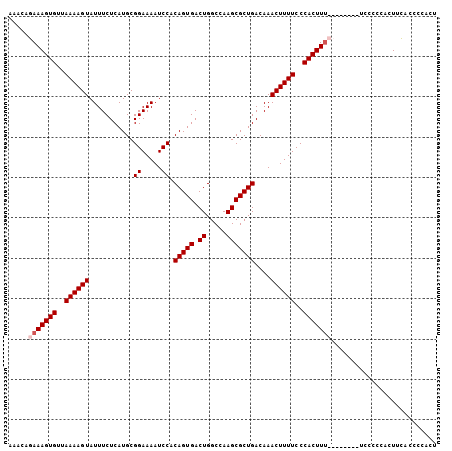
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:28 2006