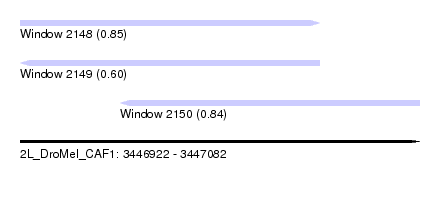
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,446,922 – 3,447,082 |
| Length | 160 |
| Max. P | 0.850858 |

| Location | 3,446,922 – 3,447,042 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -32.40 |
| Energy contribution | -32.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
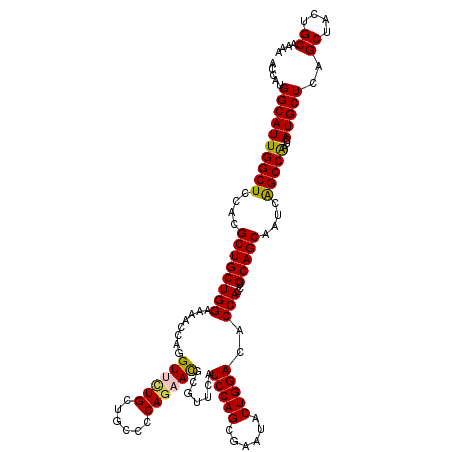
>2L_DroMel_CAF1 3446922 120 + 22407834 ACCAUGGCAUUGGCUCCACGCUGCUGGAAAACCAGGUUUUGCUGCCCCAGAACGCGUUCAUCCAGCGAGUACUGGACACCACAUGCAGCAAUCAGCCGAUCAUGCUCAGCUACUGCAAAA ..((((..(((((((....((((((((....((((..(((((((.....(((....)))...)))))))..))))...)))...)))))....)))))))))))....((....)).... ( -37.50) >DroSec_CAF1 13779 120 + 1 ACCAUGGCAUUGGCUCCACGCUGCUGGAAAACCAGGUUUUGUUGCCCCAGAACGCGUUCAUCCAGCGAAUACUGGACACCACCUGCAGCAAUCAGCCGAUCAUGCUUAGCUACUGCAAAA ..((((..(((((((....(((((.((....((((..(((((((.....(((....)))...)))))))..))))......)).)))))....)))))))))))....((....)).... ( -35.80) >DroEre_CAF1 13158 120 + 1 ACCAUGGCAUUGGCUCCACGCUGCUGGAGAACCAGGUGCUGCUGCCCCAAAACGCGUUCAUCCAGAGAAUACUGGACACCACCUGCAGCAAUCAGCCAAUCAUGCUCAGCUACUGCAAGA ..((((..(((((((....(((((.((.(((((.((.((....)).)).....).)))).(((((......))))).....)).)))))....)))))))))))....((....)).... ( -36.60) >DroYak_CAF1 944 120 + 1 AUCAUGGCAUUGGCUCCACGCUGCUGGAAAACCAGGUACUGCUGCCGCAGAAUGCGUUCAUCCAGCGAAUACUGGACACCACCUGCAGCAAUCGGCCACUGAUGCUCAGCUACUGCAAAA ....(((((((((((....((((((((....)))(((.((((....))))..........(((((......))))).)))....)))))....)))))...)))).))((....)).... ( -37.30) >consensus ACCAUGGCAUUGGCUCCACGCUGCUGGAAAACCAGGUUCUGCUGCCCCAGAACGCGUUCAUCCAGCGAAUACUGGACACCACCUGCAGCAAUCAGCCAAUCAUGCUCAGCUACUGCAAAA .....((((((((((....((((((((........((((((......)))))).......(((((......)))))..)))...)))))....)))))...)))))..((....)).... (-32.40 = -32.40 + -0.00)

| Location | 3,446,922 – 3,447,042 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -44.70 |
| Consensus MFE | -38.76 |
| Energy contribution | -38.95 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3446922 120 - 22407834 UUUUGCAGUAGCUGAGCAUGAUCGGCUGAUUGCUGCAUGUGGUGUCCAGUACUCGCUGGAUGAACGCGUUCUGGGGCAGCAAAACCUGGUUUUCCAGCAGCGUGGAGCCAAUGCCAUGGU ....(((((((((((......)))))).))))).((((.(((((((((((....)))))))...((((((((((((.(((........))))))))).))))))..))))))))...... ( -46.20) >DroSec_CAF1 13779 120 - 1 UUUUGCAGUAGCUAAGCAUGAUCGGCUGAUUGCUGCAGGUGGUGUCCAGUAUUCGCUGGAUGAACGCGUUCUGGGGCAACAAAACCUGGUUUUCCAGCAGCGUGGAGCCAAUGCCAUGGU ..(((((((((...(((.......)))..)))))))))((((((((((((....)))))))...((((((((((((.(((........))))))))).))))))........)))))... ( -43.60) >DroEre_CAF1 13158 120 - 1 UCUUGCAGUAGCUGAGCAUGAUUGGCUGAUUGCUGCAGGUGGUGUCCAGUAUUCUCUGGAUGAACGCGUUUUGGGGCAGCAGCACCUGGUUCUCCAGCAGCGUGGAGCCAAUGCCAUGGU .((((((((((.(.(((.......))).)))))))))))((((((((((......))))))...((((((((((((.(.(((...))).).)))))).))))))..)))).......... ( -45.00) >DroYak_CAF1 944 120 - 1 UUUUGCAGUAGCUGAGCAUCAGUGGCCGAUUGCUGCAGGUGGUGUCCAGUAUUCGCUGGAUGAACGCAUUCUGCGGCAGCAGUACCUGGUUUUCCAGCAGCGUGGAGCCAAUGCCAUGAU ..........((((((.(((((..((.(.(((((((((((((((((((((....)))))))..)).)).))))))))))).))..))))).)).))))..(((((........))))).. ( -44.00) >consensus UUUUGCAGUAGCUGAGCAUGAUCGGCUGAUUGCUGCAGGUGGUGUCCAGUAUUCGCUGGAUGAACGCGUUCUGGGGCAGCAAAACCUGGUUUUCCAGCAGCGUGGAGCCAAUGCCAUGGU ...((((((((.(.(((.......))).))))))))).(((((((((((......))))))...((((((((((((.(((........))))))))).))))))........)))))... (-38.76 = -38.95 + 0.19)

| Location | 3,446,962 – 3,447,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -48.00 |
| Consensus MFE | -40.14 |
| Energy contribution | -40.20 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
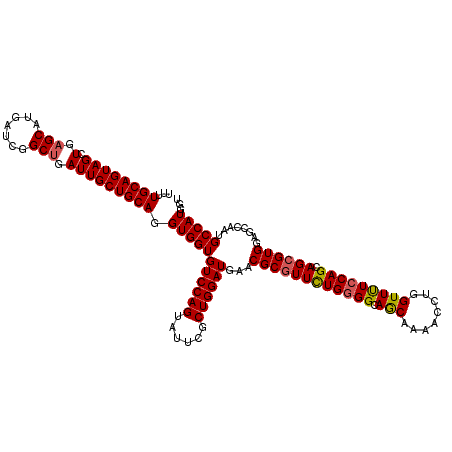
>2L_DroMel_CAF1 3446962 120 - 22407834 AGGUUGCCGAUUUCGGGACCUCCCCAGAGGAUGGCCAAGGUUUUGCAGUAGCUGAGCAUGAUCGGCUGAUUGCUGCAUGUGGUGUCCAGUACUCGCUGGAUGAACGCGUUCUGGGGCAGC (((((.(((....))))))))((((((((..((((((..((...(((((((((((......)))))).))))).))...))))(((((((....)))))))...))..)))))))).... ( -52.00) >DroSec_CAF1 13819 120 - 1 AGGUUGCCGAUUUCGGGACCUCCCCAGAGGAUGGCCAGGGUUUUGCAGUAGCUAAGCAUGAUCGGCUGAUUGCUGCAGGUGGUGUCCAGUAUUCGCUGGAUGAACGCGUUCUGGGGCAAC (((((.(((....))))))))((((((((..((((((....((((((((((...(((.......)))..))))))))))))))(((((((....)))))))...))..)))))))).... ( -48.90) >DroEre_CAF1 13198 120 - 1 AGGUUUCCGAUUUCGGGUCCUCCCCAGAGGAUGGCCAAGGUCUUGCAGUAGCUGAGCAUGAUUGGCUGAUUGCUGCAGGUGGUGUCCAGUAUUCUCUGGAUGAACGCGUUUUGGGGCAGC (((..((((....)))).)))((((((((..((((((....((((((((((.(.(((.......))).)))))))))))))))((((((......))))))...))..)))))))).... ( -46.20) >DroYak_CAF1 984 120 - 1 AGGUUGCCAAUUUCAGGACCUCCCCACAGAAUGGCCAACGUUUUGCAGUAGCUGAGCAUCAGUGGCCGAUUGCUGCAGGUGGUGUCCAGUAUUCGCUGGAUGAACGCAUUCUGCGGCAGC ..((((((.......((......)).(((((((((((....((((((((((.((.((.......)))).))))))))))))))(((((((....))))))).....))))))).)))))) ( -44.90) >consensus AGGUUGCCGAUUUCGGGACCUCCCCAGAGGAUGGCCAAGGUUUUGCAGUAGCUGAGCAUGAUCGGCUGAUUGCUGCAGGUGGUGUCCAGUAUUCGCUGGAUGAACGCGUUCUGGGGCAGC ..((((((......(((.....)))..((((((((((....((((((((((.(.(((.......))).)))))))))))))))((((((......)))))).....))))))..)))))) (-40.14 = -40.20 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:25 2006