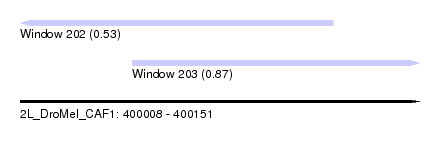
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 400,008 – 400,151 |
| Length | 143 |
| Max. P | 0.872471 |

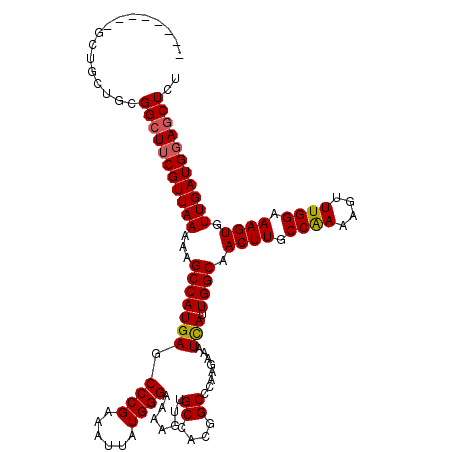
| Location | 400,008 – 400,120 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -29.61 |
| Energy contribution | -29.58 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 400008 112 - 22407834 --------GCCACUGCGGCUUCGUUAAAAAGCCAUGAGCCCGAAAUUAUGGGAAAAGUUGCCUCGGCCCAAGAAAUUAUUGGCAACUUGCCAAAAGUUUGGAAAGUGUUGAUGGAGCUCU --------((....))(((((((((((...((.....))(((((.((.(((...((((((((..................)))))))).))).)).)))))......))))))))))).. ( -30.97) >DroVir_CAF1 40774 112 - 1 --------AGCGCUGCGGCUACGUUAAAAAGCCAUGAGCCCGAAAUUAUGGGAAAAGUUGCUACGGCCCAAGAAAUCAUUGGCAACUUGCCAAAAGUUUGGAAAGUGUUGAUGGAGCUCU --------(((((((..((.((.........((((((........)))))).....)).))..))))........((((..(((.(((.((((....)))).))))))..)))).))).. ( -31.14) >DroPse_CAF1 37028 120 - 1 CUGCAGGAGCUGUUGCGGCUUCGUUAAAAUGCCAUGAGCCCGAAAUUAUGGGAAAAGUUGCCUCGGCCCAAGAAAUCAUUGGCAACUUGCCGAAAGUUUGGAAAGUGUUGAUGGAGCUCU ..((.((.((((..((((((((((....)))((((((........))))))...)))))))..))))))......((((..(((.(((.((((....)))).))))))..)))).))... ( -37.00) >DroGri_CAF1 41457 112 - 1 --------GCUGGUGUGGCUUCGUUAAAAAGCCAUGAGCCCGAAAUUAUGGGAAAAGUUGCCAUGGCUCAAGAAAUCAUUGGCAACUUGCCAAAAGUUUGGAAAGUGUUGAUGGAACUCU --------......(((((((.......)))))))(((((.......((((.(.....).)))))))))......((((..(((.(((.((((....)))).))))))..))))...... ( -32.01) >DroWil_CAF1 48682 110 - 1 ----------UGCUGUGGCUUCGUUAAAAGGCCAUGAGCCCGAAAUUAUGGGAAAAGUUGCAACAGCCCAAGAAAUCAUUGGCAACUUGCCAAAAGUUUGGAAAGUGUUGAUGGAGCUCU ----------....(((((((.......)))))))((((((((..((.((((....((....))..)))).))..))((..(((.(((.((((....)))).))))))..)))).)))). ( -31.90) >DroPer_CAF1 37517 120 - 1 CUGCAGGAGCUGUUGCGGCUUCGUUAAAAUGCCAUGAGCCCGAAAUUAUGGGAAAAGUUGCCUCGGCCCAAGAAAUCAUUGGCAACUUGCCGAAAGUUUGGAAAGUGUUGAUGGAGCUCU ..((.((.((((..((((((((((....)))((((((........))))))...)))))))..))))))......((((..(((.(((.((((....)))).))))))..)))).))... ( -37.00) >consensus ________GCUGCUGCGGCUUCGUUAAAAAGCCAUGAGCCCGAAAUUAUGGGAAAAGUUGCCACGGCCCAAGAAAUCAUUGGCAACUUGCCAAAAGUUUGGAAAGUGUUGAUGGAGCUCU ................(((((((((((...(((((((.((((......)))).......((....))........))).)))).((((.((((....)))).)))).))))))))))).. (-29.61 = -29.58 + -0.03)

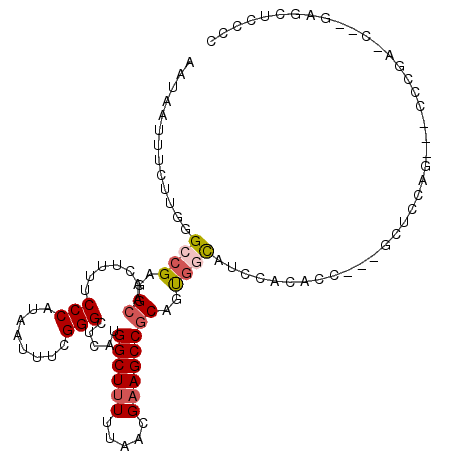
| Location | 400,048 – 400,151 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 400048 103 + 22407834 AAUAAUUUCUUGGGCCGAGGCAACUUUUCCCAUAAUUUCGGGCUCAUGGCUUUUUAACGAAGCCGCAGUGGCAUCCACACC------CGAG---CCCGAGCGCGAGCUCCCC ..........((((..(((....)))..)))).......((((((..((((((.....))))))...((((...))))...------.)))---)))((((....))))... ( -42.80) >DroVir_CAF1 40814 97 + 1 AAUGAUUUCUUGGGCCGUAGCAACUUUUCCCAUAAUUUCGGGCUCAUGGCUUUUUAACGUAGCCGCAGCGCUUGCACCACCA--ACUCCUC-------------CACUGCGC ..........((((((((.((.......(((........))).....((((.........)))))).)))...)).)))...--.......-------------........ ( -17.80) >DroEre_CAF1 23020 99 + 1 AAUAAUUUCUUGGGCCGAGGCAACUUUUCCCAUAAUUUCGGGCUCAUGGCUUUUUAACGAAGCCGCAGUGGCAUCCACACCCGAGCUCGAG---CCCGAA----------CC ..........((((..(((....)))..))))....(((((((((..((((((.....))))))...((((...))))..........)))---))))))----------.. ( -38.60) >DroYak_CAF1 15959 109 + 1 AAUAAUUUCUUGGGCCGAGGCAACUUUUCCCAUAAUUUCGGGCUCAUGGCUUUUUAACGAAGCCGCAGUGGCAUCCACACCCGAGCUCGAG---CCCGAGUGCGAGCUCCCC ........(((((((.(((....)))...........((((((((..((((((.....))))))...((((...))))....)))))))))---))))))............ ( -39.70) >DroMoj_CAF1 18873 109 + 1 AAUGAUUUCUUCGGCCGUGGCAACUUUUCCCAUAAUUUCGGGCUCAUGGCUUUUUAACGAAGCCACAGCAGCGGUCACUCCA--GCUCCACCCGCUCCU-CCUGCACUGCGC ............((((((.((.......(((........)))....(((((((.....)))))))..)).))))))......--........(((....-........))). ( -26.50) >DroAna_CAF1 37665 103 + 1 AAUGAUUUCUUGGGCCGAGGCAACUUUUCCCAUAAUUUCGGGCUCAUGGCUUUUUAACGAAGCCGCAGUGCCACCCGCA-----GCUCCAG---CACCU-CCAGAGCUCCUC ...((..(..((((..(((....)))..))))..)..))((((((..((((((.....))))))(((((((.....)))-----.))...)---)....-...))))))... ( -31.90) >consensus AAUAAUUUCUUGGGCCGAGGCAACUUUUCCCAUAAUUUCGGGCUCAUGGCUUUUUAACGAAGCCGCAGUGGCAUCCACACC___GCUCCAG___CCCGA_C__GAGCUCCCC .............((((..((.......(((........))).....((((((.....))))))))..))))........................................ (-16.09 = -16.57 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:57 2006