
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,441,243 – 3,441,353 |
| Length | 110 |
| Max. P | 0.963501 |

| Location | 3,441,243 – 3,441,353 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.11 |
| Mean single sequence MFE | -28.09 |
| Consensus MFE | -18.95 |
| Energy contribution | -20.51 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3441243 110 + 22407834 UUCAGCAUGCAUUUAUCCUUAAUGGAUUGUGCAUGUCUGCUGCCGCAGUCUUGCCACUCUUAUACCCGCCGC--CAAGACCGUUACUCCCGUAUAAGAGUUCGCUGAGCUU--C ((((((((((((..((((.....)))).))))))(.((((....)))).).....((((((((((.......--................))))))))))..))))))...--. ( -33.00) >DroAna_CAF1 34888 97 + 1 UC----UUGCUUUUAUCCUUAAUGGACUGUGCAUGU-------------CUUACCACUCUUAUACUCGUUGGUGCAUGACCGUUACGCUCGUAUAAGAGUUCGUUGAGCUUUGC ..----...........((((((((((.......))-------------).....((((((((((.(((((((.....))))..)))...)))))))))).)))))))...... ( -23.10) >DroSim_CAF1 30364 110 + 1 UUCAGCAUGCAUUUAUCCUUAAUGGAUUGUGCAUGUCUGCUGCCGCAGUCUUGCCACUCUUAUACCCGCCGC--CGAGACCGUUAUUCCCGUAUAAAAGUUCGCUGAGCUU--C ((((((((((((..((((.....)))).))))))(.((((....)))).).....(((.((((((.......--((....))........)))))).)))..))))))...--. ( -28.16) >consensus UUCAGCAUGCAUUUAUCCUUAAUGGAUUGUGCAUGUCUGCUGCCGCAGUCUUGCCACUCUUAUACCCGCCGC__CAAGACCGUUACUCCCGUAUAAGAGUUCGCUGAGCUU__C ....((((((((...(((.....)))..))))))))((((....)))).(((((.((((((((((.........................))))))))))..)).)))...... (-18.95 = -20.51 + 1.56)

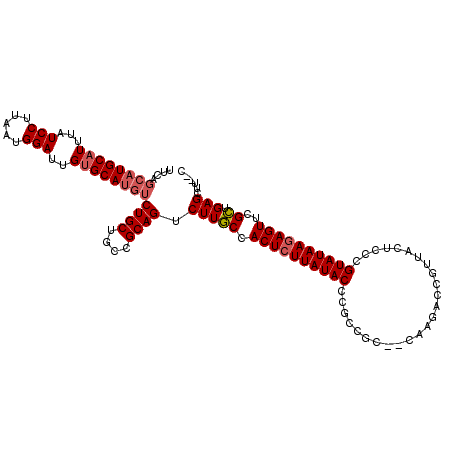

| Location | 3,441,243 – 3,441,353 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.11 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3441243 110 - 22407834 G--AAGCUCAGCGAACUCUUAUACGGGAGUAACGGUCUUG--GCGGCGGGUAUAAGAGUGGCAAGACUGCGGCAGCAGACAUGCACAAUCCAUUAAGGAUAAAUGCAUGCUGAA .--..(((..((..(((((......)))))...(((((((--.(.((..........)).)))))))))))))..(((.((((((..((((.....))))...))))))))).. ( -35.10) >DroAna_CAF1 34888 97 - 1 GCAAAGCUCAACGAACUCUUAUACGAGCGUAACGGUCAUGCACCAACGAGUAUAAGAGUGGUAAG-------------ACAUGCACAGUCCAUUAAGGAUAAAAGCAA----GA (((...((..((..((((((((((...(((...(((.....))).))).)))))))))).)).))-------------...)))...((((.....))))........----.. ( -24.50) >DroSim_CAF1 30364 110 - 1 G--AAGCUCAGCGAACUUUUAUACGGGAAUAACGGUCUCG--GCGGCGGGUAUAAGAGUGGCAAGACUGCGGCAGCAGACAUGCACAAUCCAUUAAGGAUAAAUGCAUGCUGAA .--....((((((.((((((((((.(......).(((...--..)))..))))))))))..)....((((....))))..(((((..((((.....))))...)))))))))). ( -32.20) >consensus G__AAGCUCAGCGAACUCUUAUACGGGAGUAACGGUCUUG__GCGGCGGGUAUAAGAGUGGCAAGACUGCGGCAGCAGACAUGCACAAUCCAUUAAGGAUAAAUGCAUGCUGAA ..........((..(((((((((((.......).(((.......)))..)))))))))).))....((((....)))).((((((..((((.....))))...))))))..... (-22.70 = -23.27 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:17 2006