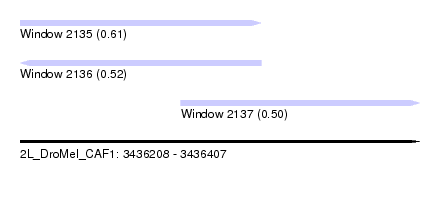
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,436,208 – 3,436,407 |
| Length | 199 |
| Max. P | 0.610815 |

| Location | 3,436,208 – 3,436,328 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -15.43 |
| Energy contribution | -15.10 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3436208 120 + 22407834 AAAAAAGAAAUAACAUUCAAUGCCAUGCAGACUGAAUGCUCCACUGCAUUUUGUUGCCUGAUAACAAUUCGCUUCGGUUACCUGGCCUUAUUUGUUACUCGUGUGUUUGCACUGGUAAUU ......(((......)))..((((((((((((.((((((......))))))....((.(((((((((...(((..(....)..))).....)))))).))).))))))))).)))))... ( -28.30) >DroSec_CAF1 25204 90 + 1 A-------------AUUCAAUGCCAUGCAGACUGAAUGCUCCACUGCAUUUUGUUGCCUGAUAACAAUUCGCCUCGGUUACCU-----------------GUGUGUUUGCACUGGUAAUU .-------------.........((.((((...((((((......))))))..)))).))...............(((((((.-----------------((((....)))).))))))) ( -22.90) >DroAna_CAF1 30148 100 + 1 A-----------ACUUUCAAUGUCAUGCAGACUGAGUGCCCCAAUGCAUUUGGUUGCCAGAUAAUCAUUUUCAACGGUUACUUUGGUUUAUU---------UGUGUUUUCAGUUGUAAUU .-----------.....((((.....((((((..(((((......)))))..)))(((((((((((.........))))).)))))).....---------..))).....))))..... ( -21.00) >DroSim_CAF1 25377 92 + 1 A-----------AUAUUCAAUGCCAAGCAGACUGAAAGCUCCACUGCAUUUUGUUGCCUGAUAACAAUUCGCCUCGGUUACCU-----------------GUGUGUUUGCACUGGUAAUU .-----------........(((((.((((((....((..((...((...((((((.....))))))...))...))....))-----------------....))))))..)))))... ( -19.00) >DroEre_CAF1 25716 107 + 1 A-----------ACAUUCAAUGUCAUGCGGACUGAAUGCUCCACUGCAUUUGGUUGUCUGAUAACAAUUUGCCUCGCUUACCUGGCUUUAUUUGUUACU--UGUGUUUGCAUUGGUAAUU .-----------....((((((.((.((((((..(((((......)))))..)))......((((((...(((..........))).....))))))..--..))).))))))))..... ( -26.80) >DroYak_CAF1 26831 107 + 1 A-----------ACAUUCAAUGUCAUGCAGACUGAAUGCUCCACUGCAUUUGGUUGCCCGAUAACAAUUUGCCUCGGUUACCUGGCUUUAUUUGUGAUU--UGUGUUUGCACUGGUAAUU .-----------........(..(((((((((..(((((......)))))..)))((.(((..((((...(((..(....)..))).....))))...)--)).)).)))).))..)... ( -25.40) >consensus A___________ACAUUCAAUGCCAUGCAGACUGAAUGCUCCACUGCAUUUGGUUGCCUGAUAACAAUUCGCCUCGGUUACCUGGCUUUAUU_________UGUGUUUGCACUGGUAAUU ....................((((((((((((.((((((......)))))).((.((((((.......)))....))).)).......................))))))).)))))... (-15.43 = -15.10 + -0.33)


| Location | 3,436,208 – 3,436,328 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -14.52 |
| Energy contribution | -16.27 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3436208 120 - 22407834 AAUUACCAGUGCAAACACACGAGUAACAAAUAAGGCCAGGUAACCGAAGCGAAUUGUUAUCAGGCAACAAAAUGCAGUGGAGCAUUCAGUCUGCAUGGCAUUGAAUGUUAUUUCUUUUUU .....(((.((((.((......))..........(((.((((((...........)))))).))).......)))).)))((((((((((((....)).))))))))))........... ( -28.70) >DroSec_CAF1 25204 90 - 1 AAUUACCAGUGCAAACACAC-----------------AGGUAACCGAGGCGAAUUGUUAUCAGGCAACAAAAUGCAGUGGAGCAUUCAGUCUGCAUGGCAUUGAAU-------------U ..(((((.(((......)))-----------------.)))))..((..(((..(((((((((((.....(((((......)))))..))))).)))))))))..)-------------) ( -20.60) >DroAna_CAF1 30148 100 - 1 AAUUACAACUGAAAACACA---------AAUAAACCAAAGUAACCGUUGAAAAUGAUUAUCUGGCAACCAAAUGCAUUGGGGCACUCAGUCUGCAUGACAUUGAAAGU-----------U ......((((.........---------......((((.(((...((((.....(.....)...))))....))).))))........(((.....)))......)))-----------) ( -11.10) >DroSim_CAF1 25377 92 - 1 AAUUACCAGUGCAAACACAC-----------------AGGUAACCGAGGCGAAUUGUUAUCAGGCAACAAAAUGCAGUGGAGCUUUCAGUCUGCUUGGCAUUGAAUAU-----------U ......((((((.......(-----------------(((((((.(((((..(((((.....(....).....)))))...)))))..)).)))))))))))).....-----------. ( -21.01) >DroEre_CAF1 25716 107 - 1 AAUUACCAAUGCAAACACA--AGUAACAAAUAAAGCCAGGUAAGCGAGGCAAAUUGUUAUCAGACAACCAAAUGCAGUGGAGCAUUCAGUCCGCAUGACAUUGAAUGU-----------U .....(((.((((......--.............(((..(....)..)))...(((((....))))).....)))).)))(((((((((((.....)))..)))))))-----------) ( -22.90) >DroYak_CAF1 26831 107 - 1 AAUUACCAGUGCAAACACA--AAUCACAAAUAAAGCCAGGUAACCGAGGCAAAUUGUUAUCGGGCAACCAAAUGCAGUGGAGCAUUCAGUCUGCAUGACAUUGAAUGU-----------U .....(((.((((......--.............(((.((...))..)))...........((....))...)))).)))(((((((((((.....)))..)))))))-----------) ( -30.10) >consensus AAUUACCAGUGCAAACACA_________AAUAAAGCCAGGUAACCGAGGCAAAUUGUUAUCAGGCAACAAAAUGCAGUGGAGCAUUCAGUCUGCAUGACAUUGAAUGU___________U .....(((.((((.........................(((((.(((......)))))))).(....)....)))).))).((((((((((.....)))..)))))))............ (-14.52 = -16.27 + 1.75)

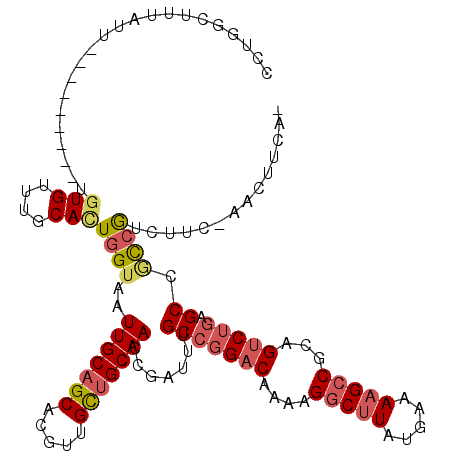
| Location | 3,436,288 – 3,436,407 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -31.03 |
| Consensus MFE | -21.84 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3436288 119 + 22407834 CCUGGCCUUAUUUGUUACUCGUGUGUUUGCACUGGUAAUUGCAGCACGCUGCUGCAAACGAUUUGCCGGACAAAAGGCUUAUAAAAAGCCGCAGUCUGAGCCAUCGUCUUCGAACUUCA- ..............(((((.((((....)))).)))))(((((((.....)))))))(((((..(((((((....(((((.....)))))...))))).)).)))))............- ( -37.40) >DroSec_CAF1 25271 102 + 1 CCU-----------------GUGUGUUUGCACUGGUAAUUGCAGCACGUUGCUGCAACCGAUUUGCCGGACAAAAGGCCUAUGAAAAGCCGCAGUCUGAGCCGCCGACUUCGAACUUCA- ...-----------------....(((((...((((..(((((((.....))))))).......(((((((....(((.........)))...))))).)).))))....)))))....- ( -28.70) >DroAna_CAF1 30217 108 + 1 CUUUGGUUUAUU---------UGUGUUUUCAGUUGUAAUUGCAGCACGUUGUUGCAACCGAUUUGCCGGACAA-AGGCUUAUGAAAAGCCCCAGCCAACGC-UUCGUCUUU-AACUUCAA ..((((((..((---------(((..............(((((((.....)))))))(((......)))))))-)(((((.....)))))..))))))...-.........-........ ( -24.70) >DroSim_CAF1 25446 102 + 1 CCU-----------------GUGUGUUUGCACUGGUAAUUGCAGCACGUUGCUGCAACCGAUUUGCCGGACAAAAGACUUAUGAAAAGCCGCAGUCUGAGCCGCCGACUUCGAACUUCA- ((.-----------------((((....)))).))...(((((((.....))))))).(((.(((.(((.....(((((..((......)).)))))...))).)))..))).......- ( -27.80) >DroEre_CAF1 25785 105 + 1 CCUGGCUUUAUUUGUUACU--UGUGUUUGCAUUGGUAAUUGCAGCGCGUUGCAGCAACCGAUUUGCCGGACAAAAGGCUUAUGAAAAGCCGCAGUCUGAGCCACCAU------------- ..((((((...((((....--..(((((((((((((..((((((....))))))..)))))..)).))))))...(((((.....)))))))))...))))))....------------- ( -33.60) >DroYak_CAF1 26900 116 + 1 CCUGGCUUUAUUUGUGAUU--UGUGUUUGCACUGGUAAUUGCAGCACGUUGUUGCAACCAAUUUGCCGGACAAAAGGCUUAUGAAAAGCCGCAGUCUGAGCCGCCGUCUUC-AACUUCG- ...(((((...((((..((--((((....).((((((((((((((.....))))))).....)))))))))))).(((((.....)))))))))...))))).........-.......- ( -34.00) >consensus CCUGGCUUUAUU_________UGUGUUUGCACUGGUAAUUGCAGCACGUUGCUGCAACCGAUUUGCCGGACAAAAGGCUUAUGAAAAGCCGCAGUCUGAGCCGCCGUCUUC_AACUUCA_ ......................(((....)))((((..(((((((.....))))))).......(((((((....(((((.....)))))...))))).)).)))).............. (-21.84 = -22.48 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:12 2006