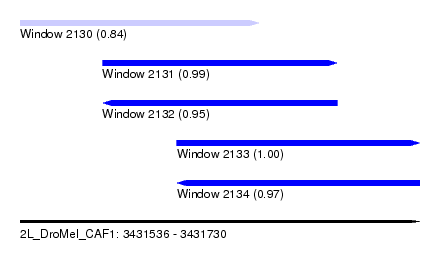
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,431,536 – 3,431,730 |
| Length | 194 |
| Max. P | 0.999928 |

| Location | 3,431,536 – 3,431,652 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.56 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -18.37 |
| Energy contribution | -18.81 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3431536 116 + 22407834 UUUAAGUUUUCAAUCGAACUGUGGUAUUUUUAAAAGGCACACACACACAC----ACCUAUUUGCAUAUGGAAAUCAUUUCUUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCC ....(((((......)))))...............(((.(((........----.....(((((((((((((..((.....)).....)))))))))))))...(((....))))))))) ( -23.70) >DroSec_CAF1 20454 111 + 1 UUUAAGUUUCCAAACGAACUGUGGUAUU-UAAAAAGGCACACAC----AC----ACAUAUUUGCAUAUGGAAAUCAUUUCUUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCC (((((((..(((.........))).)))-))))..(((.(((..----..----.(((.(((((((((((((..((.....)).....))))))))))))))))(((....))))))))) ( -26.20) >DroSim_CAF1 20536 115 + 1 UUUAAGUUUCCAAACGAACUGUGGCAUU-UAAAAAGGCACACACACACAC----ACACAUUUGCAUAUGGAAAUCAUUUCUUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCC (((((((..(((.........))).)))-))))..(((.(((........----.....(((((((((((((..((.....)).....)))))))))))))...(((....))))))))) ( -24.60) >DroEre_CAF1 20792 114 + 1 UUUAAGUUUCCAAACGAACGAUGGCAUU-UAAAAA-GCAUCCACACACAC----ACAUAUUUGCAUAUGGAAAUCAUUUCUCGUUCGAUUUCAUAUGCAAAAUACGAAUUUUCGGUGGCC (((((((..(((.........))).)))-))))..-....((((......----.....((((((((((.(((((...........)))))))))))))))...(((....))))))).. ( -23.50) >DroYak_CAF1 21921 118 + 1 UUUAAGUUUCCAAGCAAAGUGUGGUAUC-UAAAAA-GCACACACACACACAAACACUUAUUUGCAUAUGGAAAUCAUCUCUCGUUCAAUUUCAUAUGCAAAAUACGAAUUUUCGGUGGCC .............((...(((((((...-......-)).)))))...(((.........(((((((((((((.....(....).....)))))))))))))...(((....)))))))). ( -25.20) >consensus UUUAAGUUUCCAAACGAACUGUGGUAUU_UAAAAAGGCACACACACACAC____ACAUAUUUGCAUAUGGAAAUCAUUUCUUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCC ...................................(((.(((.................(((((((((((((................)))))))))))))...(((....))))))))) (-18.37 = -18.81 + 0.44)

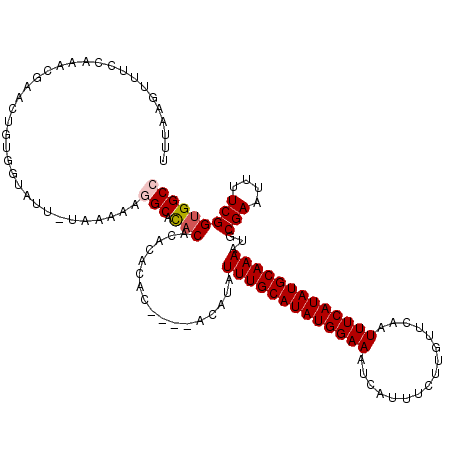
| Location | 3,431,576 – 3,431,690 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -24.62 |
| Energy contribution | -25.02 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3431576 114 + 22407834 ACACACACAC----ACCUAUUUGCAUAUGGAAAUCAUUUCUUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCCAGGACGAA-AAUGAAAU-UUGGCCAUCGCAUUGAUGGUUG ..........----(((...((((((((((((..((.....)).....))))))))))))(((((((....))((((((((((.....-.......)-))))))))))))))...))).. ( -32.00) >DroSec_CAF1 20493 110 + 1 ACAC----AC----ACAUAUUUGCAUAUGGAAAUCAUUUCUUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCCAGGACCAA-AAUGAAAU-UUGGCCAACGCAUUGAUGGUUG ....----.(----.(((.(((((((((((((..((.....)).....)))))))))))))))).)(((..((((((....((.((((-(......)-))))))....))))))..))). ( -27.60) >DroSim_CAF1 20575 114 + 1 ACACACACAC----ACACAUUUGCAUAUGGAAAUCAUUUCUUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCCAGGACCAA-AAUGAAAU-UUGGCCAACGCAUUGAUGGUUG ..........----...(((((((((((((((..((.....)).....))))))))))).((((((.......((...)).((.((((-(......)-))))))..)))))))))).... ( -26.70) >DroEre_CAF1 20830 114 + 1 CCACACACAC----ACAUAUUUGCAUAUGGAAAUCAUUUCUCGUUCGAUUUCAUAUGCAAAAUACGAAUUUUCGGUGGCCAGGACCGA-AAUGAAAU-UUGGCCAUCGCAUUGAUGGGUG .......(((----.(((.((((((((((.(((((...........)))))))))))))))...........(((((((((((.....-.......)-)))))))))).....))).))) ( -32.20) >DroYak_CAF1 21959 120 + 1 ACACACACACAAACACUUAUUUGCAUAUGGAAAUCAUCUCUCGUUCAAUUUCAUAUGCAAAAUACGAAUUUUCGGUGGCCAGGAACGAAAAUGAAAUUUUGGCCAUCGCAUUGAUGGUUG .......(((((.......(((((((((((((.....(....).....)))))))))))))...........((((((((((((............))))))))))))..))).)).... ( -29.90) >consensus ACACACACAC____ACAUAUUUGCAUAUGGAAAUCAUUUCUUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCCAGGACCAA_AAUGAAAU_UUGGCCAUCGCAUUGAUGGUUG ...................(((((((((((((................)))))))))))))...........((((((((((................))))))))))............ (-24.62 = -25.02 + 0.40)

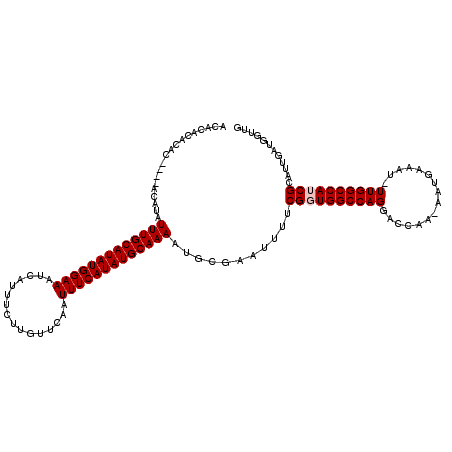
| Location | 3,431,576 – 3,431,690 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -24.11 |
| Energy contribution | -24.11 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3431576 114 - 22407834 CAACCAUCAAUGCGAUGGCCAA-AUUUCAUU-UUCGUCCUGGCCACCGAAAAUUCGCAUUUUGCAUAUGAAAUUGAACAAGAAAUGAUUUCCAUAUGCAAAUAGGU----GUGUGUGUGU ..(((((((((((((((((((.-........-.......))))))........)))))))((((((((((((((...........))))).)))))))))...)))----).))...... ( -30.19) >DroSec_CAF1 20493 110 - 1 CAACCAUCAAUGCGUUGGCCAA-AUUUCAUU-UUGGUCCUGGCCACCGAAAAUUCGCAUUUUGCAUAUGAAAUUGAACAAGAAAUGAUUUCCAUAUGCAAAUAUGU----GU----GUGU ............((.((((((.-...((...-..))...)))))).))...((.((((((((((((((((((((...........))))).)))))))))).))))----).----)).. ( -28.00) >DroSim_CAF1 20575 114 - 1 CAACCAUCAAUGCGUUGGCCAA-AUUUCAUU-UUGGUCCUGGCCACCGAAAAUUCGCAUUUUGCAUAUGAAAUUGAACAAGAAAUGAUUUCCAUAUGCAAAUGUGU----GUGUGUGUGU ..(((((.....((.((((((.-...((...-..))...)))))).))......((((((((((((((((((((...........))))).)))))))))).))))----).))).)).. ( -28.80) >DroEre_CAF1 20830 114 - 1 CACCCAUCAAUGCGAUGGCCAA-AUUUCAUU-UCGGUCCUGGCCACCGAAAAUUCGUAUUUUGCAUAUGAAAUCGAACGAGAAAUGAUUUCCAUAUGCAAAUAUGU----GUGUGUGUGG (((.(((((...((.((((((.-...((...-..))...)))))).))......(((((((.(((((((((((((.........)))))).)))))))))))))))----).))).))). ( -30.90) >DroYak_CAF1 21959 120 - 1 CAACCAUCAAUGCGAUGGCCAAAAUUUCAUUUUCGUUCCUGGCCACCGAAAAUUCGUAUUUUGCAUAUGAAAUUGAACGAGAGAUGAUUUCCAUAUGCAAAUAAGUGUUUGUGUGUGUGU ...(((((.....)))))........((((((((((((.(((...)))....(((((((.....)))))))...)))))).))))))....(((((((((((....)))))))))))... ( -29.30) >consensus CAACCAUCAAUGCGAUGGCCAA_AUUUCAUU_UUGGUCCUGGCCACCGAAAAUUCGCAUUUUGCAUAUGAAAUUGAACAAGAAAUGAUUUCCAUAUGCAAAUAUGU____GUGUGUGUGU ........(((((((((((((..................))))))........)))))))((((((((((((((...........))))).))))))))).................... (-24.11 = -24.11 + -0.00)



| Location | 3,431,612 – 3,431,730 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -33.53 |
| Energy contribution | -34.29 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.61 |
| SVM RNA-class probability | 0.999928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3431612 118 + 22407834 UUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCCAGGACGAA-AAUGAAAU-UUGGCCAUCGCAUUGAUGGUUGGCCAAAAGGCCGGCCUCAAACCAAUUUAUCACGAACGCAA ..((((.........((((((((....))))).((((((((((.....-.......)-))))))))))))((((.((((((((....)))))))))))).............)))).... ( -38.60) >DroSec_CAF1 20525 118 + 1 UUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCCAGGACCAA-AAUGAAAU-UUGGCCAACGCAUUGAUGGUUGGCCAAAAGGCCGGCCUCAAACCAAUUUAUCACGAACGCAA ..((((.........(((......(((....))).((((((((..(..-...)...)-)))))))..)))((((.((((((((....)))))))))))).............)))).... ( -35.80) >DroSim_CAF1 20611 118 + 1 UUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCCAGGACCAA-AAUGAAAU-UUGGCCAACGCAUUGAUGGUUGGCCAAAAGGCCGGCCUCAAACCAAUUUAUCACGAACGCAA ..((((.........(((......(((....))).((((((((..(..-...)...)-)))))))..)))((((.((((((((....)))))))))))).............)))).... ( -35.80) >DroEre_CAF1 20866 118 + 1 UCGUUCGAUUUCAUAUGCAAAAUACGAAUUUUCGGUGGCCAGGACCGA-AAUGAAAU-UUGGCCAUCGCAUUGAUGGGUGGCCAAGAGGACGGGCUCAAACCAAUUUAUCACGAACGCAA .(((((((((((((((.....))).....(((((((.......)))))-))))))))-(((((((((.((....)))))))))))(((......)))..............))))))... ( -33.50) >DroYak_CAF1 21999 120 + 1 UCGUUCAAUUUCAUAUGCAAAAUACGAAUUUUCGGUGGCCAGGAACGAAAAUGAAAUUUUGGCCAUCGCAUUGAUGGUUGGCCAAAAGGCCGGCCUCAAACCAAUUUAUCACGAACGCAA .(((((.........(((.......((....))(((((((((((............))))))))))))))((((.((((((((....)))))))))))).............)))))... ( -41.20) >consensus UUGUUCAAUUUCAUAUGCAAAAUGCGAAUUUUCGGUGGCCAGGACCAA_AAUGAAAU_UUGGCCAUCGCAUUGAUGGUUGGCCAAAAGGCCGGCCUCAAACCAAUUUAUCACGAACGCAA .(((((.........((((((((....))))).(((((((((................))))))))))))((((.((((((((....)))))))))))).............)))))... (-33.53 = -34.29 + 0.76)


| Location | 3,431,612 – 3,431,730 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -35.76 |
| Consensus MFE | -28.73 |
| Energy contribution | -28.81 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3431612 118 - 22407834 UUGCGUUCGUGAUAAAUUGGUUUGAGGCCGGCCUUUUGGCCAACCAUCAAUGCGAUGGCCAA-AUUUCAUU-UUCGUCCUGGCCACCGAAAAUUCGCAUUUUGCAUAUGAAAUUGAACAA ....((((((((....(((((....((((((((....))))..(((((.....)))))....-........-........)))))))))....))))(((((......))))).)))).. ( -34.60) >DroSec_CAF1 20525 118 - 1 UUGCGUUCGUGAUAAAUUGGUUUGAGGCCGGCCUUUUGGCCAACCAUCAAUGCGUUGGCCAA-AUUUCAUU-UUGGUCCUGGCCACCGAAAAUUCGCAUUUUGCAUAUGAAAUUGAACAA ....((((((((....(((((....((((((((.((((((((((((....)).)))))))))-).......-..)).)).)))))))))....))))(((((......))))).)))).. ( -38.80) >DroSim_CAF1 20611 118 - 1 UUGCGUUCGUGAUAAAUUGGUUUGAGGCCGGCCUUUUGGCCAACCAUCAAUGCGUUGGCCAA-AUUUCAUU-UUGGUCCUGGCCACCGAAAAUUCGCAUUUUGCAUAUGAAAUUGAACAA ....((((((((....(((((....((((((((.((((((((((((....)).)))))))))-).......-..)).)).)))))))))....))))(((((......))))).)))).. ( -38.80) >DroEre_CAF1 20866 118 - 1 UUGCGUUCGUGAUAAAUUGGUUUGAGCCCGUCCUCUUGGCCACCCAUCAAUGCGAUGGCCAA-AUUUCAUU-UCGGUCCUGGCCACCGAAAAUUCGUAUUUUGCAUAUGAAAUCGAACGA ...((((((.......(((((..(((......)))..(((((.(((((.....)))))((.(-(......)-).))...))))))))))...(((((((.....)))))))..)))))). ( -30.80) >DroYak_CAF1 21999 120 - 1 UUGCGUUCGUGAUAAAUUGGUUUGAGGCCGGCCUUUUGGCCAACCAUCAAUGCGAUGGCCAAAAUUUCAUUUUCGUUCCUGGCCACCGAAAAUUCGUAUUUUGCAUAUGAAAUUGAACGA ...((((((.......(((((....((((((((....))))..(((((.....)))))......................)))))))))...(((((((.....)))))))..)))))). ( -35.80) >consensus UUGCGUUCGUGAUAAAUUGGUUUGAGGCCGGCCUUUUGGCCAACCAUCAAUGCGAUGGCCAA_AUUUCAUU_UUGGUCCUGGCCACCGAAAAUUCGCAUUUUGCAUAUGAAAUUGAACAA ....(((((((((...((((((.((((....))))..))))))..))))(((((.((((((..................)))))).(((....))).....))))).......))))).. (-28.73 = -28.81 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:10 2006