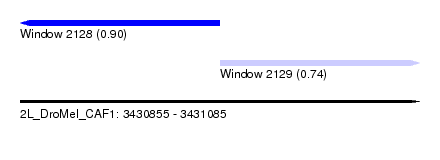
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,430,855 – 3,431,085 |
| Length | 230 |
| Max. P | 0.900418 |

| Location | 3,430,855 – 3,430,970 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
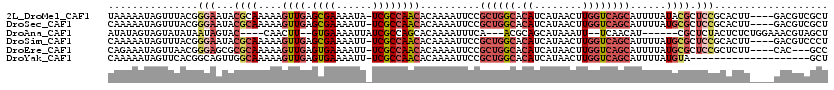
>2L_DroMel_CAF1 3430855 115 - 22407834 CAAAAUUGGUUUCAUUAAAUUACCCAAAUUAAGAUGAAAUCAUA-----UAUUUAAUACUUUCAGUUGGAAGUUAGUUGAUUAUUAGUUUAGUUAGUUGAUUACUGAGCUGAAAGUUAUC ......((((((((((.((((.....))))..))))))))))..-----........((((((((((((.(((((..((((((......))))))..))))).)).)))))))))).... ( -27.40) >DroSec_CAF1 19782 100 - 1 CAGAAUGGAUUUCAUUAAAUUACCCAAAUUAAGAUGAAAUCAUAUUUGAUAUUUAAUACUUUUAGUUGGAAGUUAGU--------------------UGAUUACUGAGCUGAAAGUUAUG ((((...(((((((((.((((.....))))..)))))))))...)))).........((((((((((((.((((...--------------------.)))).)).)))))))))).... ( -19.40) >DroSim_CAF1 19862 100 - 1 CAGAAUGGAUUUCAUUAAAUUACCCAAAUUAAGAUGAAAUCAUAUUUGAUAUUUAAUACUUUUAGUUGGAAGUUAGU--------------------UGAUUACUGAGCUGAAAGUUAUG ((((...(((((((((.((((.....))))..)))))))))...)))).........((((((((((((.((((...--------------------.)))).)).)))))))))).... ( -19.40) >consensus CAGAAUGGAUUUCAUUAAAUUACCCAAAUUAAGAUGAAAUCAUAUUUGAUAUUUAAUACUUUUAGUUGGAAGUUAGU____________________UGAUUACUGAGCUGAAAGUUAUG .......(((((((((.((((.....))))..)))))))))................((((((((((((.(((((..((((((......))))))..))))).)).)))))))))).... (-18.27 = -18.83 + 0.56)


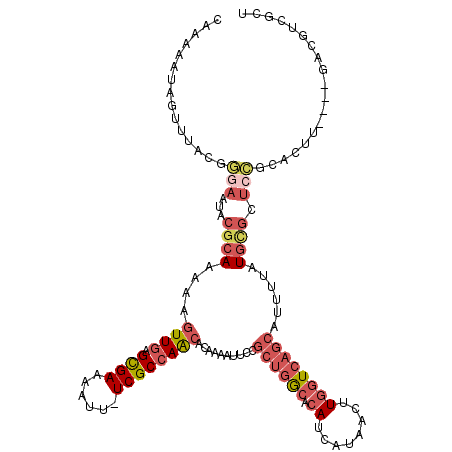
| Location | 3,430,970 – 3,431,085 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.53 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -14.57 |
| Energy contribution | -16.30 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3430970 115 + 22407834 UAAAAAUAGUUUACGGGAAUACGCAAAAAGUUGAGCGAAAAUA-UCGCCAACACAAAAUUCCGCUGGCACAUCAUAACUUGGUCAGCAUUUUAUACGCUCCGCACUU----GACGUCGCU ...............(((....((.....((((.((((.....-))))))))..........((((((.((........)))))))).........)))))((((..----...)).)). ( -25.50) >DroSec_CAF1 19882 115 + 1 CAAAAAUAGUUUACGGGAAUACGCAAAAAGUUGAGCGAAAAUU-UCGCCAACACAAAAUUCCGCUGGCACAUCAUAACUUGGUCAGCAUUUUAUGCGCUCCGCACUU----GACGUCGCU ...............(((...((((.(((((((.((((.....-))))))))..........((((((.((........))))))))..))).)))).)))((((..----...)).)). ( -29.30) >DroAna_CAF1 25635 103 + 1 AUAUAGUAGUAUAUAAUAGUAC----CAACUU--GUGAAAAUUAUCGCCAGCACAAAAUUUCA---ACGCAGCAUAAAUU--UCAACAU------CGCUCUACUCUCUGGAAACGUAGCU ....(((((........(((..----..)))(--((((((.((((.((..((...........---..)).)))))).))--)).))).------....)))))((.((....)).)).. ( -11.12) >DroSim_CAF1 19962 115 + 1 CAAAAAUAGUUUACGGGAAUACGCAAAAAGUUGAGCGAAAAUU-UCGCCAACACAAAAUUCCGCUGGCACAUCAUAACUUGGUCAGCAUUUUAUGCGCUCCGCACUU----GACGUCCCU ..............((((...((((.(((((((.((((.....-))))))))..........((((((.((........))))))))..))).))))...(((....----).)))))). ( -30.50) >DroEre_CAF1 20269 112 + 1 CAGAAAUAGUUAACGGGAGCGCGCAAAAAGUUGAGUGAAAAUU-UCGCCAACACAAAAUUCCGCUGGCACAUCAUAACUUGGUCAGCAUUUUAUGCGCUCCGCUCUU----CAC---GCC ...............((((((((.((((.((((.((((.....-))))))))..........((((((.((........)))))))).)))).))))))))......----...---... ( -35.40) >DroYak_CAF1 21396 99 + 1 CAAAAAUAGUUCACGGCAGUUGGCAAAAAGUUGAGUGAAAAUU-UCGCCAACACAAAAUUCCGCUGGCACAUCAUAACUUGGUCAGCAUUUUAUGUA--------------------GCU ..............(((.((((((...(((((.......))))-).))))))..........((((((.((........))))))))..........--------------------))) ( -23.00) >consensus CAAAAAUAGUUUACGGGAAUACGCAAAAAGUUGAGCGAAAAUU_UCGCCAACACAAAAUUCCGCUGGCACAUCAUAACUUGGUCAGCAUUUUAUGCGCUCCGCACUU____GACGUCGCU ...............(((...((((....((((.((((......))))))))..........((((((.((........))))))))......)))).)))................... (-14.57 = -16.30 + 1.73)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:05 2006