| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 392,697 – 392,790 |
| Length | 93 |
| Max. P | 0.916909 |

| Location | 392,697 – 392,790 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.13 |
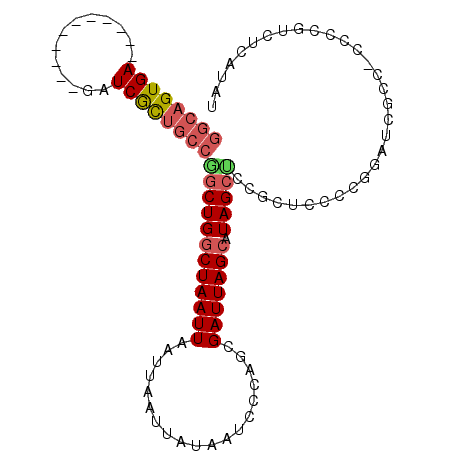
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -17.75 |
| Energy contribution | -18.41 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 392697 93 - 22407834 GGCAGUGA-----------GAUCGCUGCCGGCUGGCUAAUUAAUUAAUUAUAAUCCCAGCGAUUAGCAUAGCUCCGCUCCCCGGAUCGCCGCCCCGUCUCAUAU (((((((.-----------...)))))))(((.((((((((....)))))..((((.((((...(((...))).))))....)))).))))))........... ( -29.30) >DroSec_CAF1 8300 93 - 1 GGCAGUGA-----------GAUCGCUGCCGGCUGGCUAAUUAAUUAAUUAUAAUCCCAGCGAUUAGCAUAGCUCCGCUCCCCGGAUCGCCGCCCCGUCUCAUAU (((((((.-----------...)))))))(((.((((((((....)))))..((((.((((...(((...))).))))....)))).))))))........... ( -29.30) >DroSim_CAF1 8260 93 - 1 GGCAGUGA-----------GAUCGCUGCCGGCUGGCUAAUUAAUUAAUUAUAAUCCCAGCGAUUAGCAUAGCCCCGCUCCCCGGAUCGCCGCCCCGUCUCAUAU (((((((.-----------...)))))))(((.((((((((....)))))........((((((.....(((...))).....)))))).)))..)))...... ( -27.80) >DroEre_CAF1 15038 92 - 1 GGCAGUGA-----------GAUCGCUGCCGGCUGGCUAAUUAAUUAAUUAUAAUCCCAGCGAUUAGCAUAGCUCCGCUCCCCAGCUCGCC-CCCCGUCUCAUAC (((((((.-----------...)))))))(((.((((((((....))))........((((...(((...))).))))....)))).)))-............. ( -25.60) >DroYak_CAF1 8609 75 - 1 GGCAGUGA-----------GAUCGCUGCCGGCUGGCUAAUUAAUUAAUUAUAAUCCCAGCGAUUAGCAUAG-----------------CC-CCCCUUCUCGUAC (((((((.-----------...)))))))((((((((((((...................))))))).)))-----------------))-............. ( -25.41) >DroPer_CAF1 29732 93 - 1 ---AGUGAUCCCUAAAGCGGAUCAUGGCUAGCUGCCUAAUUAAUUAAUUAUAAUCCCACCGAUUAGCAUAGCUCCCAACUUGGGGUCUC--------CUCGCAU ---.((((((((....).)))))))(((.....)))..............(((((.....)))))((..((..(((.....)))..)).--------...)).. ( -21.70) >consensus GGCAGUGA___________GAUCGCUGCCGGCUGGCUAAUUAAUUAAUUAUAAUCCCAGCGAUUAGCAUAGCUCCGCUCCCCGGAUCGCC_CCCCGUCUCAUAU ((((((((.............))))))))((((((((((((...................))))))).)))))............................... (-17.75 = -18.41 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:53 2006