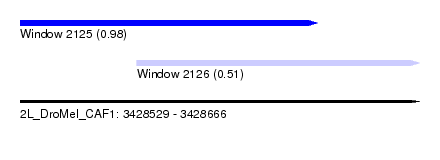
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,428,529 – 3,428,666 |
| Length | 137 |
| Max. P | 0.978742 |

| Location | 3,428,529 – 3,428,631 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -45.33 |
| Consensus MFE | -26.87 |
| Energy contribution | -27.19 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.40 |
| Mean z-score | -4.04 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
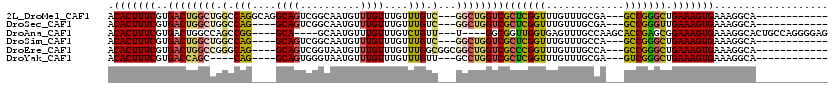
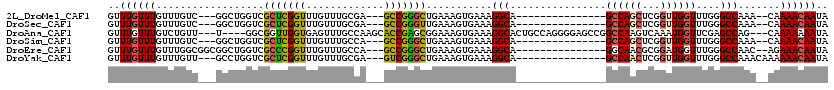
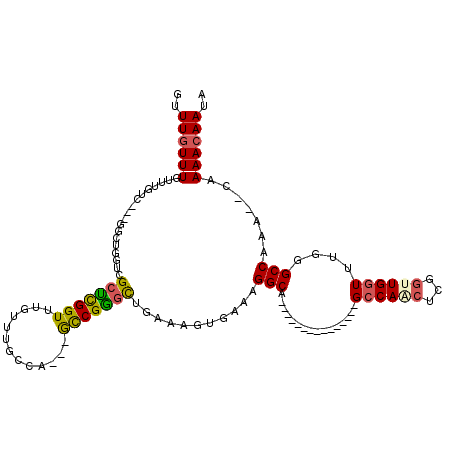
>2L_DroMel_CAF1 3428529 102 + 22407834 ACACUUUCGUGACUGGCUGGCCAGGCAGGCAGUCGGCAAUGUUUGUUUGUUUGUC---GGCUGGUCGCUCGGUUUGUUUGCGA---GCCGGGCUGAAAGUGAAAGGCA------------ .(((((((..(((..((((((.((((((((((.((....)).)))))))))))))---)))..)))((((((((((....)))---))))))).))))))).......------------ ( -53.20) >DroSec_CAF1 17471 98 + 1 ACACUUUCGUGACUGGCUGGCCAG----GCAGUCGGCAAUGUUUGUUUGUUUGUC---GGCUGGUCGCUCGGUUUGUUUGCGA---GCCGGGUUGAAAGUGAAAGGCA------------ .((((((((.(((..(((((((((----((((.((....)).)))))))...)))---)))..)))((((((((((....)))---))))))))))))))).......------------ ( -45.60) >DroAna_CAF1 19733 105 + 1 ACACUUUCGUGACUGGCCAGCCGG----GCA----GCAAUGUUUGUUUGUCUGUU---U----GGCGGUUGGUGAGUUUGCCAAGCACCGAGCGGAAAGUGAAAGGCACUGCCAGGGGAG .(((....))).((((((((.(((----(((----....)))))).)))((((((---(----((.(.(((((......))))).).))))))))).((((.....)))))))))..... ( -40.30) >DroSim_CAF1 17581 98 + 1 ACACUUUCGUGACUGGCUGGCCAG----GCAGUCGGCAAUGUUUGUUUGUUUGUC---GGCUGGUCGCUCGGUUUGUUUGCCA---GCCGGGCUGAAAGUGAAAGGCA------------ .(((((((..(((..(((((((((----((((.((....)).)))))))...)))---)))..)))((((((((.(....).)---))))))).))))))).......------------ ( -44.30) >DroEre_CAF1 17926 101 + 1 ACACUUUCGUGACUGGCCGGGCAG----GCAGUCGGUAAUGUUUGUUUGUUUGGCGGCGGCUGGUCGCCCGGUUUGUUUGCCA---GCCGGGCUGAAAGUGAAAGGCA------------ .(((((((..(((..(((((.(((----((((.(((......))).))))))).)..))))..)))((((((((.(....).)---))))))).))))))).......------------ ( -48.90) >DroYak_CAF1 19008 94 + 1 ACACUUUCGUGACCAGC----CAG----GCAGUGGGUAAUGUUUGUUUGUUUGUU---GCCUGGUCGCUCGGUUUGUUUGCGA---GUCGGGCUGAAAGUGAAAGGCA------------ .(((((((.......((----(((----((((..(((((.......)))))..))---))))))).(((((..(((....)))---..))))).))))))).......------------ ( -39.70) >consensus ACACUUUCGUGACUGGCUGGCCAG____GCAGUCGGCAAUGUUUGUUUGUUUGUC___GGCUGGUCGCUCGGUUUGUUUGCCA___GCCGGGCUGAAAGUGAAAGGCA____________ .(((((((..((((((((.(.(((....((((..........))))....))).)...))))))))(((((((.............))))))).)))))))................... (-26.87 = -27.19 + 0.31)
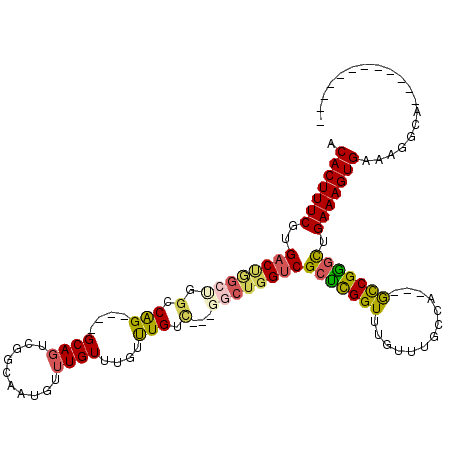
| Location | 3,428,569 – 3,428,666 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -16.97 |
| Energy contribution | -17.05 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.47 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3428569 97 + 22407834 GUUUGUUUGUUUGUC---GGCUGGUCGCUCGGUUUGUUUGCGA---GCCGGGCUGAAAGUGAAAGGCA---------------GCCAGCUCGGUUGGUUUGGGCCAAA--CAAAACAAUA ..(((((((((((..---(((((.((((((((((((....)))---)))))))(....).....))))---------------))).((((((.....))))))))))--).)))))).. ( -37.80) >DroSec_CAF1 17507 97 + 1 GUUUGUUUGUUUGUC---GGCUGGUCGCUCGGUUUGUUUGCGA---GCCGGGUUGAAAGUGAAAGGCA---------------GCCAGCUCGGUUGGUUUGGGCCAAA--CAAAACAAUA ..(((((((((((..---(((((.((((((((((((....)))---)))))).)))...(....).))---------------))).((((((.....))))))))))--).)))))).. ( -35.60) >DroAna_CAF1 19765 110 + 1 GUUUGUUUGUCUGUU---U----GGCGGUUGGUGAGUUUGCCAAGCACCGAGCGGAAAGUGAAAGGCACUGCCAGGGGAGCCGGCCAAGUCAAAUGGUUCGAGCCAG---CAAAAAAAUA (((((((((((((((---(----((.(.(((((......))))).).))))))))).((((.....))))(((.((....))))))))).)))))(((....)))..---.......... ( -36.50) >DroSim_CAF1 17617 97 + 1 GUUUGUUUGUUUGUC---GGCUGGUCGCUCGGUUUGUUUGCCA---GCCGGGCUGAAAGUGAAAGGCA---------------GCCAGCUCGGUUGGUUUGGGCCAAA--CAAAACAAUA ..((((((((((...---(((.....))).(((..(...((((---(((((((((...((.....)).---------------..))))))))))))).)..))))))--).)))))).. ( -36.50) >DroEre_CAF1 17962 100 + 1 GUUUGUUUGUUUGGCGGCGGCUGGUCGCCCGGUUUGUUUGCCA---GCCGGGCUGAAAGUGAAAGGCA---------------GGCAACGCGGAUGGUUUGGGCCAAC--AGAAACAAUA ..((((((.(((((((((((....)))))(.(((((((((((.---(((...(.......)...))).---------------))))).)))))).).....))))).--).)))))).. ( -36.60) >DroYak_CAF1 19040 99 + 1 GUUUGUUUGUUUGUU---GCCUGGUCGCUCGGUUUGUUUGCGA---GUCGGGCUGAAAGUGAAAGGCA---------------GCCAACUCGGUUGGUUUGGGCCAAACAAAAAACAAUA ((((.((((((((..---(((..(.((((((..(((....)))---..))))).............((---------------(((.....)))))).)..))))))))))))))).... ( -34.40) >consensus GUUUGUUUGUUUGUC___GGCUGGUCGCUCGGUUUGUUUGCCA___GCCGGGCUGAAAGUGAAAGGCA_______________GCCAACUCGGUUGGUUUGGGCCAAA__CAAAACAAUA ..((((((..................(((((((.............)))))))...........(((................((((((...))))))....))).......)))))).. (-16.97 = -17.05 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:02 2006