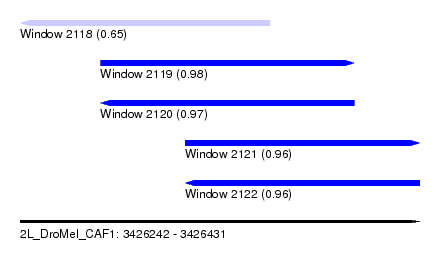
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,426,242 – 3,426,431 |
| Length | 189 |
| Max. P | 0.982879 |

| Location | 3,426,242 – 3,426,360 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.27 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -17.88 |
| Energy contribution | -19.68 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3426242 118 - 22407834 GACAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAACAAUAGUACUGGCACAUACAUGCAUACAUGUAUGUAGCACAGUGUAGAUAUGUAUGUAGUUAUAUGACGGAUAUAGCA .(((.(((..(((....)))((((((((.....((....((....))....))(((((((((....))))))))).)))).)))).....))).))).(((((((.....))))))). ( -35.70) >DroSec_CAF1 15120 106 - 1 --CAAACAAACUCAAAAGAGGCGCGUGCCGAAAGCAAAAACAAUAGUACUGGCACAUACAU------AUGUAUGUAGCACAGUGUAGAUAU----GUAGUUAUAUGACGGAUAUAGCA --...(((..(((....)))((((((((.....((....((....))....))((((((..------..)))))).)))).)))).....)----)).(((((((.....))))))). ( -24.70) >DroSim_CAF1 15292 106 - 1 --CAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAACAAUAGUACUGGCACAUACAU------AUGUAUGUAGCACAGUGUAGAUAU----AUAGUUAUAUGACGGAUAUAGCA --........(((....)))((..((((((................(((((((((((((..------..)))))).)).)))))...((((----(....)))))..))).))).)). ( -28.30) >DroEre_CAF1 15528 102 - 1 --CAAACAAACUCCAAAGAGGCGCGUGCCGAUAGCAAAAACAAUAGUGCUGCCCCAU----------AUGUAUGUAGCGCAGUGUAGAUAU----GUAUUUCUAUGGCGGAUACAGCA --........(((....)))((..((((((...............(((((((.....----------......)))))))...(((((...----.....)))))..))).))).)). ( -22.60) >DroYak_CAF1 16577 106 - 1 --CAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAACAAUAGUGCUGCCACAUUUGG------AUGUAUGUAGCACAGUGUAGAUAU----GUUUUUGUAUGGCGGAUAAAGCA --........(((....)))((.....(((...(((((((((...(((((((.((((....------))))..))))))).((....)).)----))))))))....))).....)). ( -32.10) >consensus __CAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAACAAUAGUACUGGCACAUACAU______AUGUAUGUAGCACAGUGUAGAUAU____GUAGUUAUAUGACGGAUAUAGCA ..........(((....)))(((((((((....)...................((((((((......)))))))).)))).)))).............(((((((.....))))))). (-17.88 = -19.68 + 1.80)

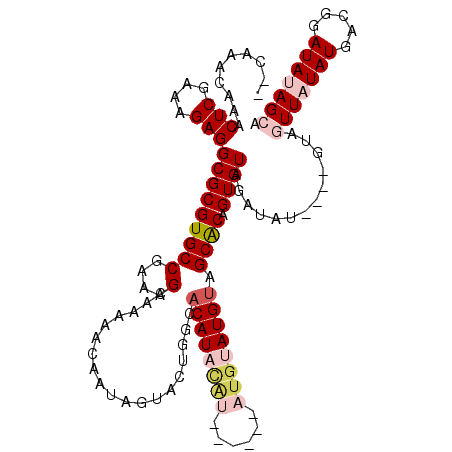
| Location | 3,426,280 – 3,426,400 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.79 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -26.08 |
| Energy contribution | -28.48 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3426280 120 + 22407834 GUGCUACAUACAUGUAUGCAUGUAUGUGCCAGUACUAUUGUUUUUGCUUUCGGCACGCGCCUCUUUCGAGUUUGUUUGUCGUCUGCCUUCGAUUUUUAACAAACAAACUCAAAAGUGCGA ((((((((((((((....)))))))))((.((.((....)).)).))....))))).(((..((((.((((((((((((.(((.......))).....)))))))))))).)))).))). ( -41.10) >DroSec_CAF1 15154 111 + 1 GUGCUACAUACAU------AUGUAUGUGCCAGUACUAUUGUUUUUGCUUUCGGCACGCGCCUCUUUUGAGUUUGUUUG---UCUGCCUUCGAUUUUUAACAAACAAACGCAGAAGUGCGA (((((((((((..------..))))))((.((.((....)).)).))....))))).(((..((((((.(((((((((---(................)))))))))).)))))).))). ( -34.49) >DroSim_CAF1 15326 111 + 1 GUGCUACAUACAU------AUGUAUGUGCCAGUACUAUUGUUUUUGCUUUCGGCACGCGCCUCUUUCGAGUUUGUUUG---UCUGCCUUCGAUUUUUAACAAACAAACGCAGAAGUGCGA (((((((((((..------..))))))((.((.((....)).)).))....)))))((((.(((.....(((((((((---(................))))))))))..))).)))).. ( -30.89) >DroEre_CAF1 15562 107 + 1 GCGCUACAUACAU----------AUGGGGCAGCACUAUUGUUUUUGCUAUCGGCACGCGCCUCUUUGGAGUUUGUUUG---UCUGCCUUCGAUUUUUAACAAACAAACUGCGAAGUGCGA (((((........----------..(((((.((((....))...(((.....))).))))))).(((.((((((((((---(................))))))))))).)))))))).. ( -32.19) >DroYak_CAF1 16611 111 + 1 GUGCUACAUACAU------CCAAAUGUGGCAGCACUAUUGUUUUUGCUUUCGGCACGCGCCUCUUUCGAGUUUGUUUG---UCUGCCUUCGAUUUUUAGCAAACAAACUCAGAAGUGCGA .((((((((....------....))))))))(((((...............(((....)))......(((((((((((---.(((...........))))))))))))))...))))).. ( -36.10) >consensus GUGCUACAUACAU______AUGUAUGUGCCAGUACUAUUGUUUUUGCUUUCGGCACGCGCCUCUUUCGAGUUUGUUUG___UCUGCCUUCGAUUUUUAACAAACAAACUCAGAAGUGCGA ((((((((((((((....)))))))))(..((((..(....)..))))..)))))).(((..((((.(((((((((((...((.......)).......))))))))))).)))).))). (-26.08 = -28.48 + 2.40)

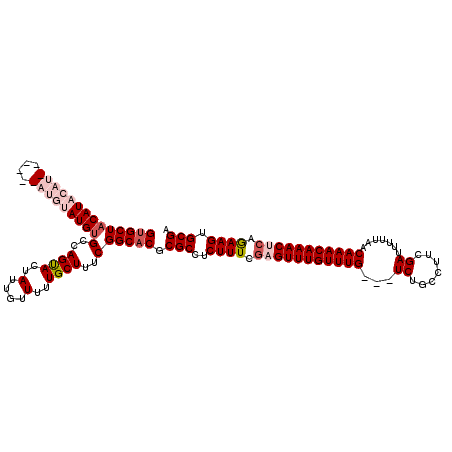
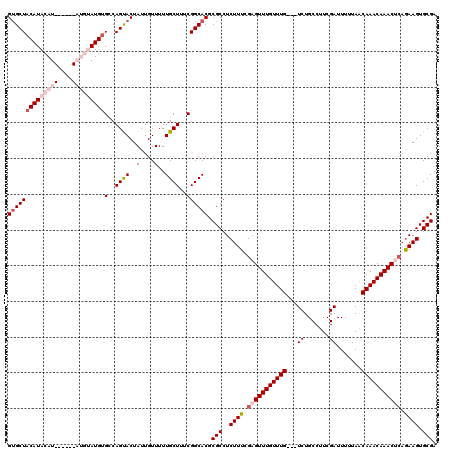
| Location | 3,426,280 – 3,426,400 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.79 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -24.80 |
| Energy contribution | -25.80 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3426280 120 - 22407834 UCGCACUUUUGAGUUUGUUUGUUAAAAAUCGAAGGCAGACGACAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAACAAUAGUACUGGCACAUACAUGCAUACAUGUAUGUAGCAC ..((.((((((((((((((((((......(....).....))))))))))))))))))..))((((((((...((..........))..)))))).((((((((....)))))))))).. ( -44.30) >DroSec_CAF1 15154 111 - 1 UCGCACUUCUGCGUUUGUUUGUUAAAAAUCGAAGGCAGA---CAAACAAACUCAAAAGAGGCGCGUGCCGAAAGCAAAAACAAUAGUACUGGCACAUACAU------AUGUAUGUAGCAC ..((.(((((..(((((((((((..............))---))))))))).....))))).))((((.....((....((....))....))((((((..------..)))))).)))) ( -30.54) >DroSim_CAF1 15326 111 - 1 UCGCACUUCUGCGUUUGUUUGUUAAAAAUCGAAGGCAGA---CAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAACAAUAGUACUGGCACAUACAU------AUGUAUGUAGCAC ..((((...((((((((((((((..........))))))---)))))...(((....)))))).))))(....).................((((((((..------..)))))).)).. ( -33.30) >DroEre_CAF1 15562 107 - 1 UCGCACUUCGCAGUUUGUUUGUUAAAAAUCGAAGGCAGA---CAAACAAACUCCAAAGAGGCGCGUGCCGAUAGCAAAAACAAUAGUGCUGCCCCAU----------AUGUAUGUAGCGC ..((((..(((.(((((((((((..........))))))---)))))...(((....)))))).)))).................(((((((.....----------......))))))) ( -29.70) >DroYak_CAF1 16611 111 - 1 UCGCACUUCUGAGUUUGUUUGCUAAAAAUCGAAGGCAGA---CAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAACAAUAGUGCUGCCACAUUUGG------AUGUAUGUAGCAC ..((((......(((((((((((..........))))))---)))))...(((....)))....))))(....)...........(((((((.((((....------))))..))))))) ( -36.40) >consensus UCGCACUUCUGAGUUUGUUUGUUAAAAAUCGAAGGCAGA___CAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAACAAUAGUACUGGCACAUACAU______AUGUAUGUAGCAC ..((.((((((((((((((((........(....).......))))))))))))...)))).))(((((....)...................((((((((......)))))))).)))) (-24.80 = -25.80 + 1.00)

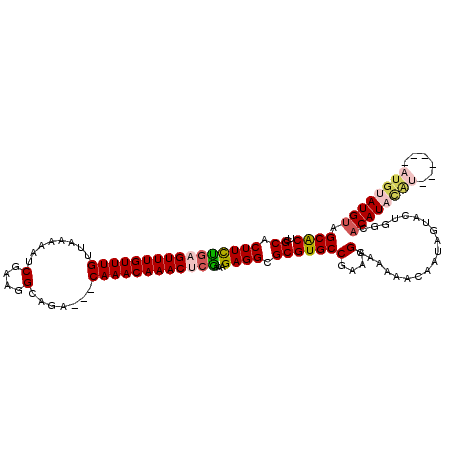
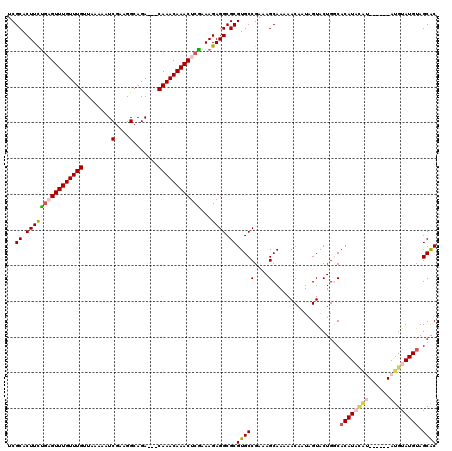
| Location | 3,426,320 – 3,426,431 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.03 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -24.16 |
| Energy contribution | -24.84 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3426320 111 + 22407834 UUUUUGCUUUCGGCACGCGCCUCUUUCGAGUUUGUUUGUCGUCUGCCUUCGAUUUUUAACAAACAAACUCAAAAGUGCGAAAUGCAGACUGAUUAACGCAGAAGCAAACAC--------- ..(((((((((.(((..(((..((((.((((((((((((.(((.......))).....)))))))))))).)))).)))...))).))(((.......))))))))))...--------- ( -33.10) >DroSec_CAF1 15188 113 + 1 UUUUUGCUUUCGGCACGCGCCUCUUUUGAGUUUGUUUG---UCUGCCUUCGAUUUUUAACAAACAAACGCAGAAGUGCGAAAUGCAGACUGAUAAACGCAGAAGCAAACACAAA----CA ..((((((((.(((....))).....((.(((((((.(---(((((.((((((((((.............)))))).))))..)))))).))))))).))))))))))......----.. ( -32.32) >DroSim_CAF1 15360 113 + 1 UUUUUGCUUUCGGCACGCGCCUCUUUCGAGUUUGUUUG---UCUGCCUUCGAUUUUUAACAAACAAACGCAGAAGUGCGAAAUGCAGACUGAUAAACGCAGAAGCAAACACAAA----CA ..((((((((.(((....)))......(.(((((((.(---(((((.((((((((((.............)))))).))))..)))))).))))))).).))))))))......----.. ( -31.72) >DroEre_CAF1 15592 109 + 1 UUUUUGCUAUCGGCACGCGCCUCUUUGGAGUUUGUUUG---UCUGCCUUCGAUUUUUAACAAACAAACUGCGAAGUGCGAAAUGCAGAGUGGUAAACGCAAAUGCAAG--------CACA ..((((((((..(((..(((..(((((.((((((((((---(................))))))))))).))))).)))...)))...)))))))).((....))...--------.... ( -34.29) >DroYak_CAF1 16645 117 + 1 UUUUUGCUUUCGGCACGCGCCUCUUUCGAGUUUGUUUG---UCUGCCUUCGAUUUUUAGCAAACAAACUCAGAAGUGCGAAAUGCAGAGUGAUAAACGCAAAUGCAAAUGCAAACACACA ..(((((..((.(((..(((..(((..(((((((((((---.(((...........))))))))))))))..))).)))...))).))((.....)))))))(((....)))........ ( -33.80) >consensus UUUUUGCUUUCGGCACGCGCCUCUUUCGAGUUUGUUUG___UCUGCCUUCGAUUUUUAACAAACAAACUCAGAAGUGCGAAAUGCAGACUGAUAAACGCAGAAGCAAACACAAA____CA ..(((((..((.(((..(((..((((.(((((((((((...((.......)).......))))))))))).)))).)))...))).))(((.......)))..)))))............ (-24.16 = -24.84 + 0.68)

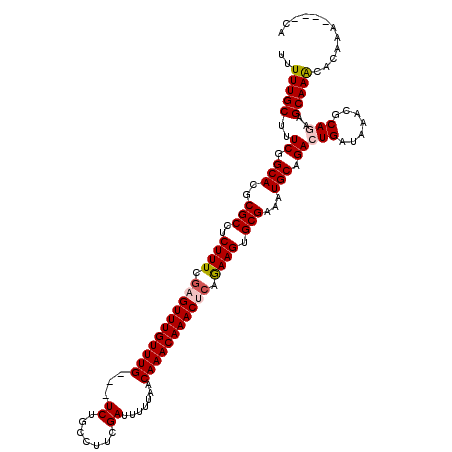
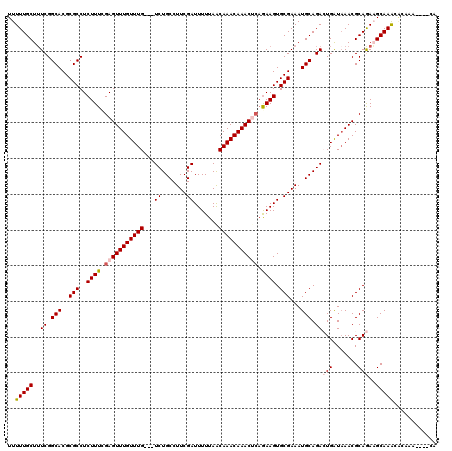
| Location | 3,426,320 – 3,426,431 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.03 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -25.56 |
| Energy contribution | -25.48 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
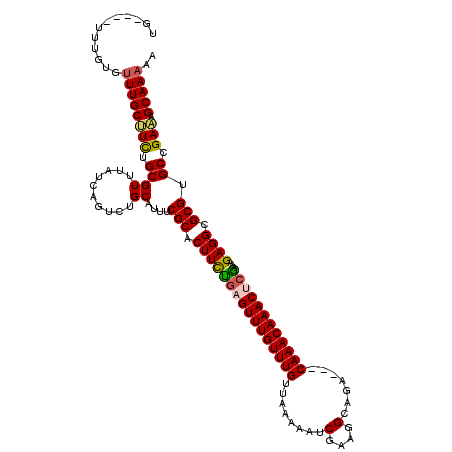
>2L_DroMel_CAF1 3426320 111 - 22407834 ---------GUGUUUGCUUCUGCGUUAAUCAGUCUGCAUUUCGCACUUUUGAGUUUGUUUGUUAAAAAUCGAAGGCAGACGACAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAA ---------...((((((((((.......)))((.((((..(((.((((((((((((((((((......(....).....))))))))))))))))))..))).)))).))))))))).. ( -38.20) >DroSec_CAF1 15188 113 - 1 UG----UUUGUGUUUGCUUCUGCGUUUAUCAGUCUGCAUUUCGCACUUCUGCGUUUGUUUGUUAAAAAUCGAAGGCAGA---CAAACAAACUCAAAAGAGGCGCGUGCCGAAAGCAAAAA ..----.(((((..(((....)))..)).)))..(((.((((((((...((((((((((((((..........))))))---)))))...(((....)))))).))).)))))))).... ( -29.70) >DroSim_CAF1 15360 113 - 1 UG----UUUGUGUUUGCUUCUGCGUUUAUCAGUCUGCAUUUCGCACUUCUGCGUUUGUUUGUUAAAAAUCGAAGGCAGA---CAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAA ..----.(((((..(((....)))..)).)))..(((.((((((((...((((((((((((((..........))))))---)))))...(((....)))))).))).)))))))).... ( -33.40) >DroEre_CAF1 15592 109 - 1 UGUG--------CUUGCAUUUGCGUUUACCACUCUGCAUUUCGCACUUCGCAGUUUGUUUGUUAAAAAUCGAAGGCAGA---CAAACAAACUCCAAAGAGGCGCGUGCCGAUAGCAAAAA .(((--------..(((....)))..))).....(((...((((((..(((.(((((((((((..........))))))---)))))...(((....)))))).))).)))..))).... ( -29.50) >DroYak_CAF1 16645 117 - 1 UGUGUGUUUGCAUUUGCAUUUGCGUUUAUCACUCUGCAUUUCGCACUUCUGAGUUUGUUUGCUAAAAAUCGAAGGCAGA---CAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAA .(((((..((((........))))..)).)))..(((.((((((((......(((((((((((..........))))))---)))))...(((....)))....))).)))))))).... ( -37.90) >consensus UG____UUUGUGUUUGCUUCUGCGUUUAUCAGUCUGCAUUUCGCACUUCUGAGUUUGUUUGUUAAAAAUCGAAGGCAGA___CAAACAAACUCGAAAGAGGCGCGUGCCGAAAGCAAAAA ............((((((((.((((..........))....(((.((((((((((((((((........(....).......))))))))))))...)))).))).)).))).))))).. (-25.56 = -25.48 + -0.08)

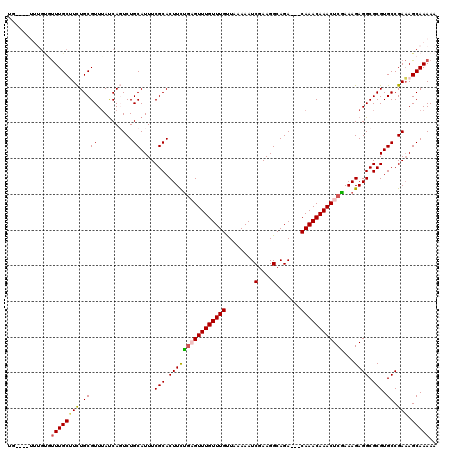
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:58 2006