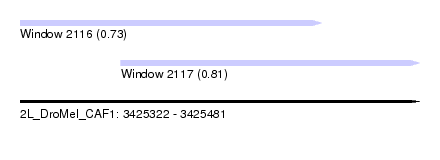
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,425,322 – 3,425,481 |
| Length | 159 |
| Max. P | 0.806246 |

| Location | 3,425,322 – 3,425,442 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.44 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -30.57 |
| Energy contribution | -31.32 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
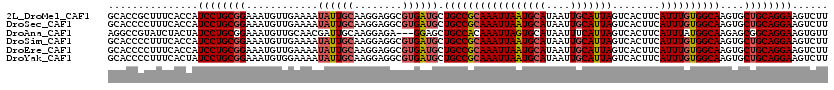
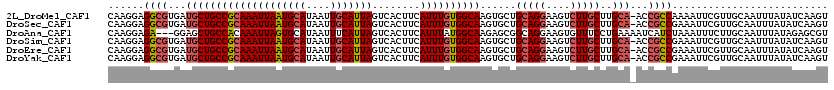
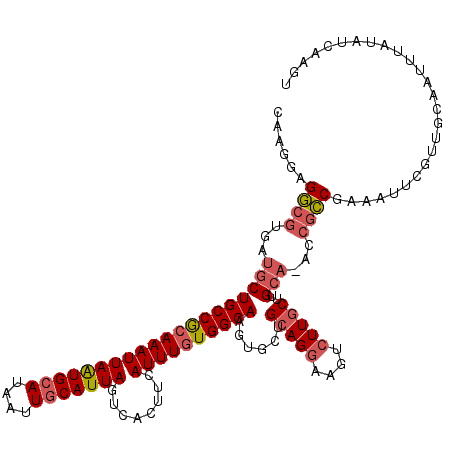
>2L_DroMel_CAF1 3425322 120 + 22407834 GCACCGCUUUCACCAUCCUGCGGAAAUGUUGAAAAUAUUGCAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUU ((((.((..(((((.(((((((...((((.....))))))).)))).).)))).))(((((((((((((((((....)))))))........)))))))))).))))............. ( -37.00) >DroSec_CAF1 14163 120 + 1 GCACCCCUUUCACCAUCCUGCGGAAAUGUUGAAAAUAUUGCAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUU ...............((((((((............(((..(........)..))).(((((((((((((((((....)))))))........))))))))))....))))))))...... ( -35.10) >DroAna_CAF1 16636 117 + 1 AGGCCGUAUCUACUAUCCUGCGGAAAUGUUGCAACGAUUGCAAGGAGA---GGAGCUGCCACAAAUUAGUGCAUAAUUUCAUUAGUCACUUCAUUUAUGGCAAGAGCGGCAGGAAGUGUU ..........((((.((((((.....(.(((((.....))))).)...---...(((((((.((((.((((..((((...))))..))))..)))).))))...))).)))))))))).. ( -29.50) >DroSim_CAF1 14337 120 + 1 GCACCCCUUUCACCAUCCUGCGGAAAUGUUGAAAAUAUUGCAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUU ...............((((((((............(((..(........)..))).(((((((((((((((((....)))))))........))))))))))....))))))))...... ( -35.10) >DroEre_CAF1 14392 120 + 1 GCACCCCUUUCACCAUCCUGCGGAAAUGUUGAAAAUAUUGCAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUU ...............((((((((............(((..(........)..))).(((((((((((((((((....)))))))........))))))))))....))))))))...... ( -35.10) >DroYak_CAF1 15633 120 + 1 GCACCCCUUUCACUAUCCUGCGGAAAUGUGGAAAAUAUUGCAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUU ...........(((.((((((((...(((((....(((..(........)..)))...)))))...(((((((....)))))))(((((.......))))).....)))))))))))... ( -37.10) >consensus GCACCCCUUUCACCAUCCUGCGGAAAUGUUGAAAAUAUUGCAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUU ...............((((((((............((((((........)))))).(((((((((((((((((....)))))))........))))))))))....))))))))...... (-30.57 = -31.32 + 0.75)
| Location | 3,425,362 – 3,425,481 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.23 |
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -29.35 |
| Energy contribution | -30.13 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
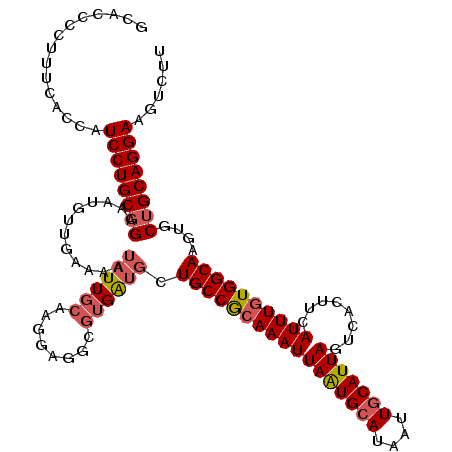
>2L_DroMel_CAF1 3425362 119 + 22407834 CAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUUGCUUGCA-ACCGCCAAAAUUCGUUGCAAUUUAUAUCAAGU (((.((((((.(.((((((((((((((((((((....)))))))........))))))))))......(((((....)))))..)))-.))))).....)).)))............... ( -36.60) >DroSec_CAF1 14203 119 + 1 CAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUUGCUUGCA-ACCGCCGAAAUUCGUUGCAAUUUAUAUCAAGU (((.((((((.(.((((((((((((((((((((....)))))))........))))))))))......(((((....)))))..)))-.))))).....)).)))............... ( -36.60) >DroAna_CAF1 16676 117 + 1 CAAGGAGA---GGAGCUGCCACAAAUUAGUGCAUAAUUUCAUUAGUCACUUCAUUUAUGGCAAGAGCGGCAGGAAGUGUUUCCUGAAAAUCAUCUAAAUUUCUUGCAAUUUAUAGAGCGU (((((((.---(((..(((((.((((.((((..((((...))))..))))..)))).))))).((....((((((....))))))....)).)))...)))))))............... ( -24.90) >DroSim_CAF1 14377 119 + 1 CAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUUGCUUGCA-ACCGCCGAAAUUCGUUGCAAUUUAUAUCAAGU (((.((((((.(.((((((((((((((((((((....)))))))........))))))))))......(((((....)))))..)))-.))))).....)).)))............... ( -36.60) >DroEre_CAF1 14432 119 + 1 CAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUUGCUUGCA-ACCGCCGAAAUUCGUUGCAAUUUAUAUCAAGU (((.((((((.(.((((((((((((((((((((....)))))))........))))))))))......(((((....)))))..)))-.))))).....)).)))............... ( -36.60) >DroYak_CAF1 15673 119 + 1 CAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUUGCUUGCA-ACCGCCGAAAUUCGUUGCAAUUUAUAUCAAGU (((.((((((.(.((((((((((((((((((((....)))))))........))))))))))......(((((....)))))..)))-.))))).....)).)))............... ( -36.60) >consensus CAAGGAGGCGUGAUGCUGCCGCAAAUUAAUGCAUAAUUGCAUUAGUCACUUCAUUUGUGGCAAGUGCUGCAGGAAGUCUUGCUUGCA_ACCGCCGAAAUUCGUUGCAAUUUAUAUCAAGU ......((((...((((((((((((((((((((....)))))))........))))))))))......(((((....)))))..)))...)))).......................... (-29.35 = -30.13 + 0.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:53 2006