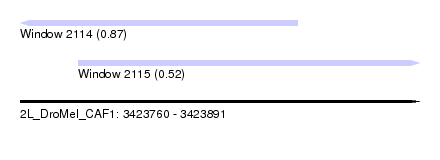
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,423,760 – 3,423,891 |
| Length | 131 |
| Max. P | 0.868701 |

| Location | 3,423,760 – 3,423,851 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 83.59 |
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -13.99 |
| Energy contribution | -13.59 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3423760 91 - 22407834 CUGGCCCACAU----ACGCUGUUUAUAAAUAAAGCACAUCUUU-UUUACGGCAACUGCA--UGUGAGAGGGCUGAAAAAAAAAAAACAAAAAAACAAA ..(((((....----..(((............))).......(-((((((((....)).--))))))))))))......................... ( -18.20) >DroSec_CAF1 12640 78 - 1 CUGGCCCACAU----ACGCUGUUUAUAAAUAAAGCACAUCUUU-UUUACGGCAACUGCA--UGUGAGAGGGCUAAAAA-------------AAGUAAA .((((((....----..(((............))).......(-((((((((....)).--)))))))))))))....-------------....... ( -19.20) >DroSim_CAF1 12817 78 - 1 CUGGCCCACAU----ACGCUGUUUAUAAAUAAAGCACAUCUUU-UUUACGGCAACUGCA--UGUGAGAGGGCUAAAAA-------------AAGUAAA .((((((....----..(((............))).......(-((((((((....)).--)))))))))))))....-------------....... ( -19.20) >DroEre_CAF1 12888 77 - 1 CUGGCCCACAU----ACGCUGUUUAUAAAUAAAGAACAUCUUU-UUUACGGCAACUGCA--UGUGGGAGGGCUGAAAA--------------AAUAAA ..(((((.(((----(.((.(((.......((((.....))))-........))).)).--))))...))))).....--------------...... ( -16.66) >DroYak_CAF1 14089 81 - 1 CUGGCCCACAUCUGUGCGCUGUUUAUAAAUAC---ACAUCUUUUUUUGCGGCAAGUGCAUAUGUGAGAGGGCUGAGAA--------------AAUAAA ..(((((.(((.((((((((..(..((((...---.........))))..)..)))))))).)))...))))).....--------------...... ( -20.60) >consensus CUGGCCCACAU____ACGCUGUUUAUAAAUAAAGCACAUCUUU_UUUACGGCAACUGCA__UGUGAGAGGGCUGAAAA_____________AAAUAAA .((((((..........(((((.........................)))))....((....))....))))))........................ (-13.99 = -13.59 + -0.40)


| Location | 3,423,779 – 3,423,891 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -19.18 |
| Energy contribution | -20.57 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3423779 112 + 22407834 UUUUUCAGCCCUCUCACA---UGCAGUUGCCGUAAA-AAAGAUGUGCUUUAUUUAUAAACAGCGU----AUGUGGGCCAGUCCAAAAAGGGAUGUCGGAUAGUGAGUGGGACGACUGGAC ...((((((((.((((((---(((.((((..(((((-((((.....)))).)))))...))))))----)))))))((.((((......))))...)).........)))....))))). ( -29.30) >DroSec_CAF1 12647 107 + 1 -UUUUUAGCCCUCUCACA---UGCAGUUGCCGUAAA-AAAGAUGUGCUUUAUUUAUAAACAGCGU----AUGUGGGCCAGUCCGAAAAGGGAUGUCGGAUAG----UGGGACGGCUGGAC -..(((((((..((((((---(((.((((..(((((-((((.....)))).)))))...))))))----)))))))(((((((((.........))))))..----)))...))))))). ( -34.30) >DroAna_CAF1 15235 99 + 1 -UUUUCCGCUUUCUGAACCGCUGCAGUUGGUGUAAAAAAAUAAACGACUUAUUUAUAAAUAGCGU----AUGUGGGGCAAUCCUAAAAAGGAUAAAG----------------GAUGAGC -......((((((((..(((((((.((((...((...((((((.....)))))).))..))))))----).))))..).(((((....)))))...)----------------)).)))) ( -19.60) >DroSim_CAF1 12824 107 + 1 -UUUUUAGCCCUCUCACA---UGCAGUUGCCGUAAA-AAAGAUGUGCUUUAUUUAUAAACAGCGU----AUGUGGGCCAGUCCGAAAAGGGAUGUCGGAUAG----UGGGACGGCUGGAC -..(((((((..((((((---(((.((((..(((((-((((.....)))).)))))...))))))----)))))))(((((((((.........))))))..----)))...))))))). ( -34.30) >DroEre_CAF1 12894 107 + 1 -UUUUCAGCCCUCCCACA---UGCAGUUGCCGUAAA-AAAGAUGUUCUUUAUUUAUAAACAGCGU----AUGUGGGCCAGUCCGAAAAGGGAUGUCGGAUAG----UGGGACGGCUGGAC -..(((((((..((((((---(((.((((..(((((-((((.....)))).)))))...))))))----)))))))(((((((((.........))))))..----)))...))))))). ( -40.20) >DroYak_CAF1 14095 111 + 1 -UUCUCAGCCCUCUCACA-UAUGCACUUGCCGCAAAAAAAGAUGU---GUAUUUAUAAACAGCGCACAGAUGUGGGCCAGUCCGAAAAGGGAUGUCGGCUAG----UGGGAUGGCUGGAC -.((.(((((.((((((.-..........(((((........(((---(..((.......))..))))..)))))(((.((((......))))...)))..)----))))).))))))). ( -35.90) >consensus _UUUUCAGCCCUCUCACA___UGCAGUUGCCGUAAA_AAAGAUGUGCUUUAUUUAUAAACAGCGU____AUGUGGGCCAGUCCGAAAAGGGAUGUCGGAUAG____UGGGACGGCUGGAC ...(((((((..((((((.......((((..(((((.((((.....)))).)))))...)))).......))))))((.((((......))))...))..............))))))). (-19.18 = -20.57 + 1.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:51 2006