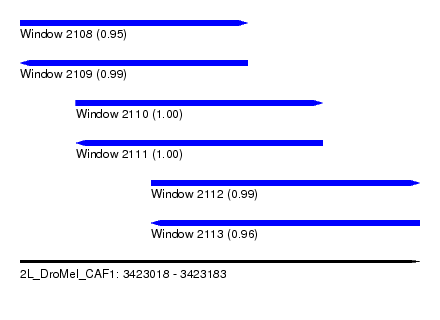
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,423,018 – 3,423,183 |
| Length | 165 |
| Max. P | 0.997787 |

| Location | 3,423,018 – 3,423,112 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 89.89 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -25.98 |
| Energy contribution | -25.60 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3423018 94 + 22407834 GCCGUAUAAGAGUCGCGUUCGAACUUCGAACGCAACGUGCUUGCUGCUCGUCUACGAGUUCGAACCGAACAACAACGGCAUCGAAUCGAACUCA ...(((........(((((((.....))))))).(((.((.....)).))).)))((((((((..(((.(......)...)))..)))))))). ( -31.50) >DroSec_CAF1 11894 94 + 1 GUCGUAUAAGAGUCGCGUUCGAACUUCGAACGCAACGUGCUCGAUGCUCGUCUUCGAGUUCGAAGCGAACAACAACGGUAUUGAAUCGAACUCG .........((((.(((((((.....)))))))..(((..((((.(((((....))))))))).)))........((((.....)))).)))). ( -31.20) >DroSim_CAF1 12081 94 + 1 GUCGUAUAAGAGUCGCGUUCGAACUUCGAACGCAACAUGCUGGACGUUCGUCUUCGAGUUCGAAGCGAACAACAACGGUAUUGAAUCGAACUCG .........((((((((((((.....)))))))...((((((...(((((.(((((....)))))))))).....))))))......).)))). ( -34.40) >DroEre_CAF1 12129 94 + 1 GUCGUAUAAGAGUCGCGUUCGAACUUCGAACGCAACAUGUUGGACGCUCGUCUUCGAGUUCGAACCGAACCACUUCGGCAUUGCACCGAACUCG .........((((((.(((((((((.((((..(((....)))(((....))))))))))))))))))).....(((((.......)))))))). ( -29.90) >consensus GUCGUAUAAGAGUCGCGUUCGAACUUCGAACGCAACAUGCUGGACGCUCGUCUUCGAGUUCGAACCGAACAACAACGGCAUUGAAUCGAACUCG .........((((((((((((.....)))))))..(((((((.....(((....)))(((((...))))).....))))).))....).)))). (-25.98 = -25.60 + -0.38)

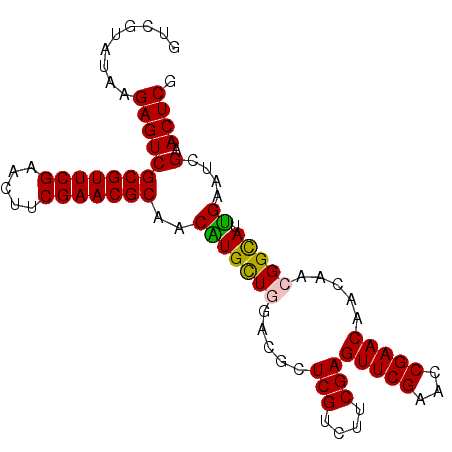
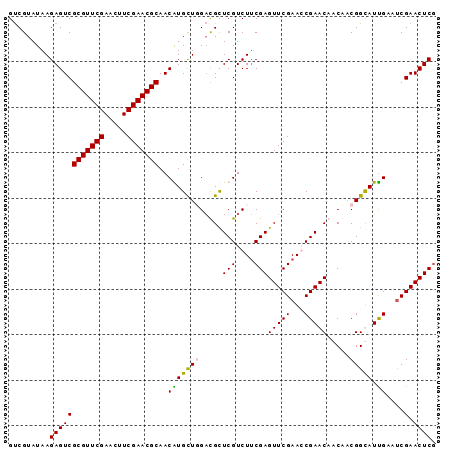
| Location | 3,423,018 – 3,423,112 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 89.89 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -30.10 |
| Energy contribution | -29.98 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3423018 94 - 22407834 UGAGUUCGAUUCGAUGCCGUUGUUGUUCGGUUCGAACUCGUAGACGAGCAGCAAGCACGUUGCGUUCGAAGUUCGAACGCGACUCUUAUACGGC ...............((((((((((((((...((....))....))))))))).....((((((((((.....))))))))))......))))) ( -36.80) >DroSec_CAF1 11894 94 - 1 CGAGUUCGAUUCAAUACCGUUGUUGUUCGCUUCGAACUCGAAGACGAGCAUCGAGCACGUUGCGUUCGAAGUUCGAACGCGACUCUUAUACGAC ((.((((((..((((...)))).(((((((((((....))))).))))))))))))..((((((((((.....)))))))))).......)).. ( -38.20) >DroSim_CAF1 12081 94 - 1 CGAGUUCGAUUCAAUACCGUUGUUGUUCGCUUCGAACUCGAAGACGAACGUCCAGCAUGUUGCGUUCGAAGUUCGAACGCGACUCUUAUACGAC .....(((..........((((.(((((((((((....))))).))))))..))))..((((((((((.....)))))))))).......))). ( -36.20) >DroEre_CAF1 12129 94 - 1 CGAGUUCGGUGCAAUGCCGAAGUGGUUCGGUUCGAACUCGAAGACGAGCGUCCAACAUGUUGCGUUCGAAGUUCGAACGCGACUCUUAUACGAC (((((((((......((((((....)))))))))))))))..(((....)))......((((((((((.....))))))))))........... ( -36.40) >consensus CGAGUUCGAUUCAAUACCGUUGUUGUUCGCUUCGAACUCGAAGACGAGCAUCCAGCACGUUGCGUUCGAAGUUCGAACGCGACUCUUAUACGAC (((((((((......((((........)))))))))))))..................((((((((((.....))))))))))........... (-30.10 = -29.98 + -0.13)

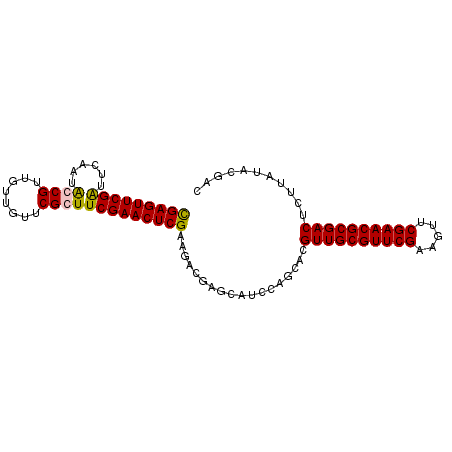
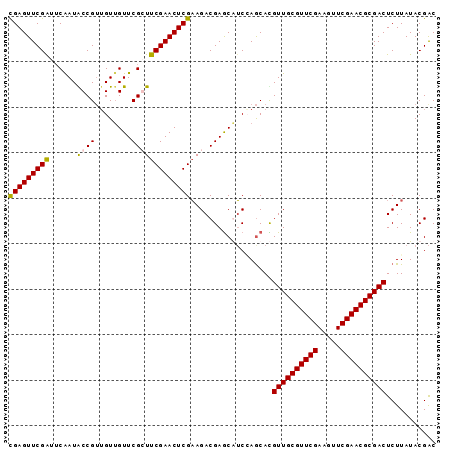
| Location | 3,423,041 – 3,423,143 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.66 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.82 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3423041 102 + 22407834 CUUCGAACGCAACGUGCUUGCUGCUCGUCUACGAGUUCGAACCGAACAACAACGGCAUCGAAUCGAACUCAAAGAUAAAAAGUUGUAAGGGAAU---------UUCUGUUA ........((((((.((.....)).)((((..((((((((..(((.(......)...)))..))))))))..)))).....)))))((.((...---------..)).)). ( -23.50) >DroSec_CAF1 11917 102 + 1 CUUCGAACGCAACGUGCUCGAUGCUCGUCUUCGAGUUCGAAGCGAACAACAACGGUAUUGAAUCGAACUCGAAGAUAAGAAGUUGUAACAGAAU---------UUCUGUUA ((((....(((.((....)).)))..((((((((((((((..(((((.......)).)))..))))))))))))))..))))...((((((...---------..)))))) ( -36.00) >DroSim_CAF1 12104 102 + 1 CUUCGAACGCAACAUGCUGGACGUUCGUCUUCGAGUUCGAAGCGAACAACAACGGUAUUGAAUCGAACUCGAAGAUAAGAAGUUGUAACAGAAU---------UUCUGUUA (((((((((...(.....)..)))))((((((((((((((..(((((.......)).)))..))))))))))))))..))))...((((((...---------..)))))) ( -36.70) >DroEre_CAF1 12152 111 + 1 CUUCGAACGCAACAUGUUGGACGCUCGUCUUCGAGUUCGAACCGAACCACUUCGGCAUUGCACCGAACUCGAAGAUAACAAGUUUUAACAGAACUUUUUCUAGUUCUGUUA ..((.((((.....)))).)).....(((((((((((((..(((((....)))))........))))))))))))).........((((((((((......)))))))))) ( -36.50) >consensus CUUCGAACGCAACAUGCUGGACGCUCGUCUUCGAGUUCGAACCGAACAACAACGGCAUUGAAUCGAACUCGAAGAUAAGAAGUUGUAACAGAAU_________UUCUGUUA ........(((((..((.....))..((((((((((((((..(((.(......)...)))..)))))))))))))).....)))))(((((((..........))))))). (-27.33 = -27.82 + 0.50)

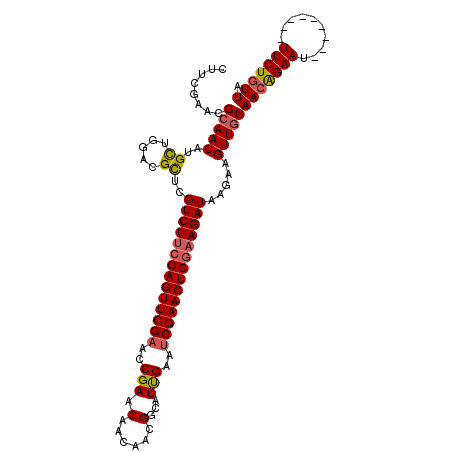
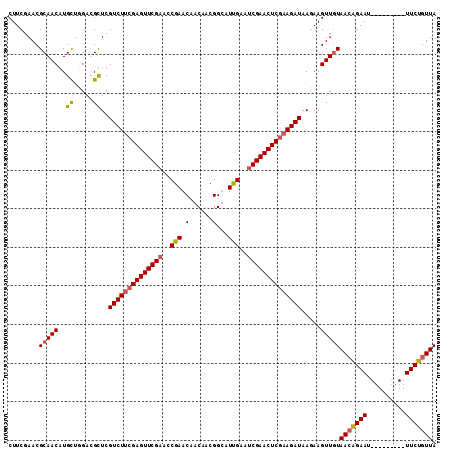
| Location | 3,423,041 – 3,423,143 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.66 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -28.46 |
| Energy contribution | -29.65 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3423041 102 - 22407834 UAACAGAA---------AUUCCCUUACAACUUUUUAUCUUUGAGUUCGAUUCGAUGCCGUUGUUGUUCGGUUCGAACUCGUAGACGAGCAGCAAGCACGUUGCGUUCGAAG ........---------...................(((.(((((((((..(((.((....))...)))..))))))))).)))(((((.((((.....)))))))))... ( -28.10) >DroSec_CAF1 11917 102 - 1 UAACAGAA---------AUUCUGUUACAACUUCUUAUCUUCGAGUUCGAUUCAAUACCGUUGUUGUUCGCUUCGAACUCGAAGACGAGCAUCGAGCACGUUGCGUUCGAAG ((((((..---------...))))))......((..(((((((((((((..(((((....)))))......)))))))))))))..))..((((((.......)))))).. ( -35.90) >DroSim_CAF1 12104 102 - 1 UAACAGAA---------AUUCUGUUACAACUUCUUAUCUUCGAGUUCGAUUCAAUACCGUUGUUGUUCGCUUCGAACUCGAAGACGAACGUCCAGCAUGUUGCGUUCGAAG ((((((..---------...))))))...((((...(((((((((((((..(((((....)))))......))))))))))))).((((((..........)))))))))) ( -35.50) >DroEre_CAF1 12152 111 - 1 UAACAGAACUAGAAAAAGUUCUGUUAAAACUUGUUAUCUUCGAGUUCGGUGCAAUGCCGAAGUGGUUCGGUUCGAACUCGAAGACGAGCGUCCAACAUGUUGCGUUCGAAG ((((((((((......))))))))))..........(((((((((((((......((((((....)))))))))))))))))))((((((..(((....)))))))))... ( -44.70) >consensus UAACAGAA_________AUUCUGUUACAACUUCUUAUCUUCGAGUUCGAUUCAAUACCGUUGUUGUUCGCUUCGAACUCGAAGACGAGCAUCCAGCACGUUGCGUUCGAAG ((((((((..........))))))))..........(((((((((((((......((((........)))))))))))))))))(((((((..........)))))))... (-28.46 = -29.65 + 1.19)

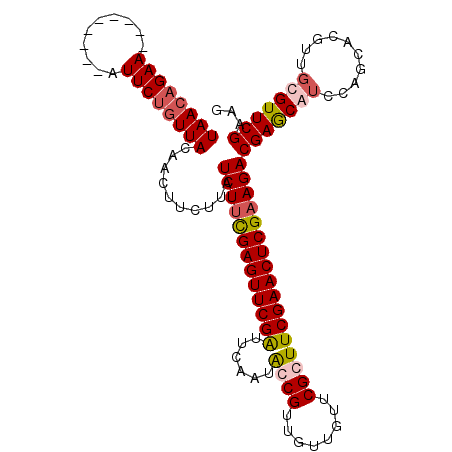
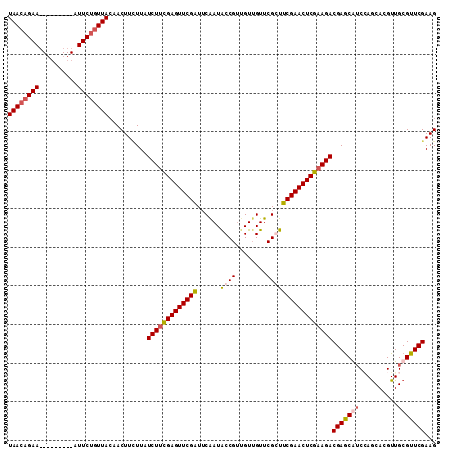
| Location | 3,423,072 – 3,423,183 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.63 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -21.16 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3423072 111 + 22407834 CGAGUUCGAACCGAACAACAACGGCAUCGAAUCGAACUCAAAGAUAAAAAGUUGUAAGGGAAU---------UUCUGUUAUUUGUCUAAUUGGAAUAGAAAUUGGAAUUUAUCUAAUUUA .((((((((..(((.(......)...)))..))))))))..(((((((.....((((.((...---------..)).))))...(((((((........))))))).)))))))...... ( -25.90) >DroSec_CAF1 11948 111 + 1 CGAGUUCGAAGCGAACAACAACGGUAUUGAAUCGAACUCGAAGAUAAGAAGUUGUAACAGAAU---------UUCUGUUAUUUGUCUAAUUGGAAUAGAAAUUGAAAUUGAACAGAUUUA (((((((((..(((((.......)).)))..)))))))))..............((((((...---------..))))))(((((.(((((..(((....)))..))))).))))).... ( -25.10) >DroSim_CAF1 12135 111 + 1 CGAGUUCGAAGCGAACAACAACGGUAUUGAAUCGAACUCGAAGAUAAGAAGUUGUAACAGAAU---------UUCUGUUAUUUGUCCAAUUGGAAUAUAAAUUGGAAUUGAACUAAUUUA (((((((((..(((((.......)).)))..))))))))).........(((((((((((...---------..)))))))...(((((((........)))))))....))))...... ( -30.10) >DroEre_CAF1 12183 120 + 1 CGAGUUCGAACCGAACCACUUCGGCAUUGCACCGAACUCGAAGAUAACAAGUUUUAACAGAACUUUUUCUAGUUCUGUUAUUUGUCCAAUUGGAAUAGCAGUUGGAAUCGAACUUAUUUA ((((((((..(((((....)))))........))))))))........((((((((((((((((......))))))))))....((((((((......))))))))...))))))..... ( -38.40) >DroYak_CAF1 13342 120 + 1 CGAGUUCGAAGCGAACCACUCCGGCAUUGAAUCAAACUCGAAGAUAAAAAGUUUUAACAGAACUUUUUCUAGUUCUGUUAUCUGUCUAAUUGGAAUGUAAAUUGGAAUUGAACUAAGUUA ((((((.((..(((.((.....))..)))..)).)))))).........(((((((((((((((......))))))))))....(((((((........)))))))...)))))...... ( -29.50) >consensus CGAGUUCGAAGCGAACAACAACGGCAUUGAAUCGAACUCGAAGAUAAAAAGUUGUAACAGAAU_________UUCUGUUAUUUGUCUAAUUGGAAUAGAAAUUGGAAUUGAACUAAUUUA (((((((((..(((............)))..)))))))))..............((((((((..........))))))))....(((((((........))))))).............. (-21.16 = -21.44 + 0.28)

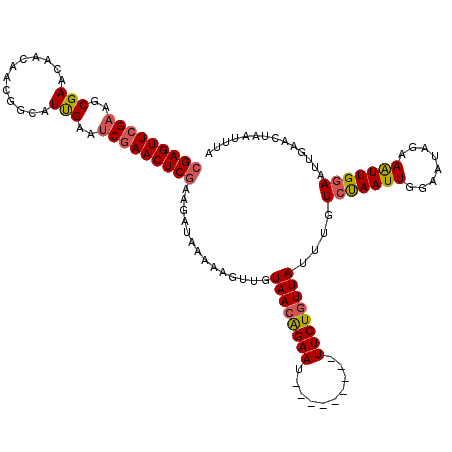
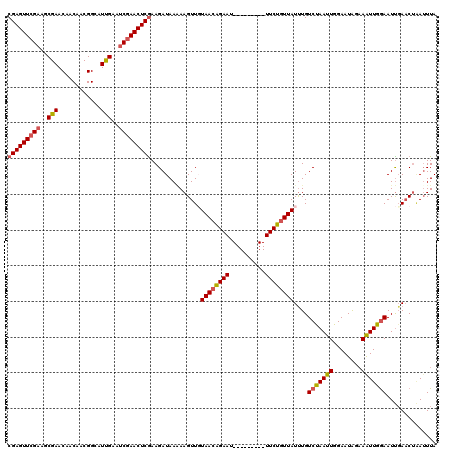
| Location | 3,423,072 – 3,423,183 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.63 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -16.90 |
| Energy contribution | -16.58 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3423072 111 - 22407834 UAAAUUAGAUAAAUUCCAAUUUCUAUUCCAAUUAGACAAAUAACAGAA---------AUUCCCUUACAACUUUUUAUCUUUGAGUUCGAUUCGAUGCCGUUGUUGUUCGGUUCGAACUCG ......(((((((....(((((((....................))))---------)))............)))))))..((((((((..(((.((....))...)))..)))))))). ( -19.34) >DroSec_CAF1 11948 111 - 1 UAAAUCUGUUCAAUUUCAAUUUCUAUUCCAAUUAGACAAAUAACAGAA---------AUUCUGUUACAACUUCUUAUCUUCGAGUUCGAUUCAAUACCGUUGUUGUUCGCUUCGAACUCG ......(((..((((..(((....)))..))))..)))..((((((..---------...))))))..............(((((((((..(((((....)))))......))))))))) ( -21.10) >DroSim_CAF1 12135 111 - 1 UAAAUUAGUUCAAUUCCAAUUUAUAUUCCAAUUGGACAAAUAACAGAA---------AUUCUGUUACAACUUCUUAUCUUCGAGUUCGAUUCAAUACCGUUGUUGUUCGCUUCGAACUCG .......((((((((..(((....)))..))))))))...((((((..---------...))))))..............(((((((((..(((((....)))))......))))))))) ( -25.40) >DroEre_CAF1 12183 120 - 1 UAAAUAAGUUCGAUUCCAACUGCUAUUCCAAUUGGACAAAUAACAGAACUAGAAAAAGUUCUGUUAAAACUUGUUAUCUUCGAGUUCGGUGCAAUGCCGAAGUGGUUCGGUUCGAACUCG ......(((((((..(((((..((..(((....)))....((((((((((......))))))))))..(((((.......)))))(((((.....)))))))..))).)).))))))).. ( -39.30) >DroYak_CAF1 13342 120 - 1 UAACUUAGUUCAAUUCCAAUUUACAUUCCAAUUAGACAGAUAACAGAACUAGAAAAAGUUCUGUUAAAACUUUUUAUCUUCGAGUUUGAUUCAAUGCCGGAGUGGUUCGCUUCGAACUCG .......((..((((..(((....)))..))))..))...((((((((((......))))))))))..............(((((((((..(...(((.....)))..)..))))))))) ( -26.00) >consensus UAAAUUAGUUCAAUUCCAAUUUCUAUUCCAAUUAGACAAAUAACAGAA_________AUUCUGUUACAACUUCUUAUCUUCGAGUUCGAUUCAAUGCCGUUGUUGUUCGCUUCGAACUCG ........................................((((((((..........))))))))..............(((((((((......((.......)).....))))))))) (-16.90 = -16.58 + -0.32)

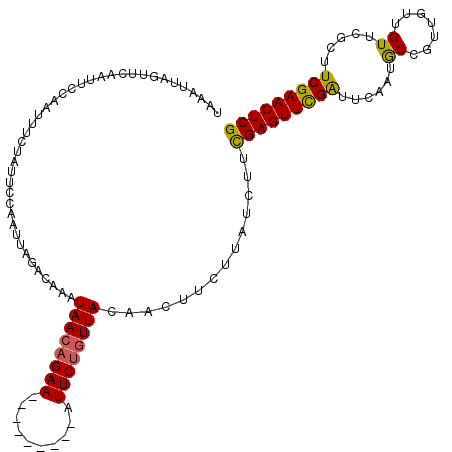

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:49 2006