| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,416,522 – 3,416,621 |
| Length | 99 |
| Max. P | 0.992898 |

| Location | 3,416,522 – 3,416,621 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.07 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -15.97 |
| Energy contribution | -17.17 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
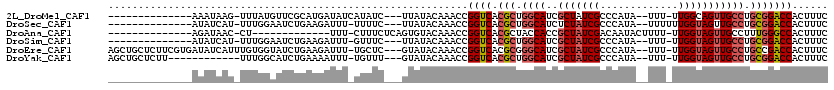
>2L_DroMel_CAF1 3416522 99 + 22407834 GAAAGUGGUCCGCAGGCAACUGCCAA-AAA--UAUGGGCGAUAGCGAUGCCAGCGUGACCGGUUUGUAUAA---GAUAUGAUAUCAUGCGAACAUAAA-CUUAUUU-------------- ..(((((((((((.((((....(((.-...--..)))((....))..)))).))).)))).((((((((..---((((...))))))))))))....)-)))....-------------- ( -31.10) >DroSec_CAF1 5459 99 + 1 GAAAGUGGUCCGCAGGCAACUACCAAAAAA--UAUGGGCGAUAGAGAUGCCAGCGUGACCGGUUUGUAUAA---GAAAA-AAAUCUUCAGAUUCCAAA-AUGAUAU-------------- .(((.((((((((.((((.((.(((.....--..))).......)).)))).))).))))).)))((((..---.....-.((((....)))).....-...))))-------------- ( -22.79) >DroAna_CAF1 7899 90 + 1 GAAAGUGGCCCAAAGGCAACUACCAA-AAAAGUAUUGUCGAUAGCGGUGGUAGCGUGACCGGUUUGUACACUGAGAAAG-AAA-------------AG-GUUAUCU-------------- ...(((.(((....))).))).....-.....(((..(((....)))..)))..((((((..(((...(.....)...)-)).-------------.)-)))))..-------------- ( -16.90) >DroSim_CAF1 5688 98 + 1 GAAAGUGGUCCGCAGGCAACUACCAA-AAA--UAUGGGCGAUAGCGAUGCCAGCGUGACCGGUUUGUAUAA---GAAAC-AAAUCUUCAGAUUCCAAA-AUGAUAU-------------- (((...(((((((.((((....(((.-...--..)))((....))..)))).))).))))(((((((....---...))-))))))))..........-.......-------------- ( -28.30) >DroEre_CAF1 5601 113 + 1 GAAAGUGGUCGGCAGGCAACUACCAA-AAA--UAUGGGCGAUAGCGAUGCCCGCGUGACCGGUUUGUAUAC---GAGCA-AAAUCUUCAGAUACCACAAAUGAUAUCACGAAGAGCAGCU .(((.((((((((.((((....(((.-...--..)))((....))..)))).)).)))))).)))......---.(((.-...(((((.((((.((....)).))))..)))))...))) ( -32.40) >DroYak_CAF1 6089 101 + 1 GAAAGUGGUCCGCAGGCAACUACCAA-AAA--UAUGGGCGAUAGCGAUGCCAGCGUGACCGGUUUGUAUAC---AAACA-AAAUUUUCAGAUGCCAAA------------AAGAGCAGCU (((((((((((((.((((....(((.-...--..)))((....))..)))).))).)))).(((((....)---)))).-..))))))((.(((....------------....))).)) ( -32.00) >consensus GAAAGUGGUCCGCAGGCAACUACCAA_AAA__UAUGGGCGAUAGCGAUGCCAGCGUGACCGGUUUGUAUAA___GAAAA_AAAUCUUCAGAUUCCAAA_AUGAUAU______________ .(((.((((((((.((((....(((.........)))((....))..)))).))).))))).)))....................................................... (-15.97 = -17.17 + 1.20)

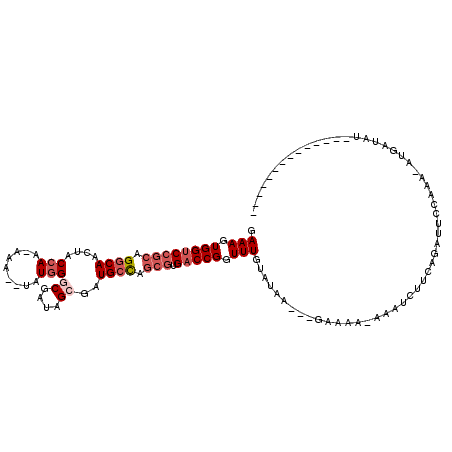
| Location | 3,416,522 – 3,416,621 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.07 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -18.73 |
| Energy contribution | -19.45 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3416522 99 - 22407834 --------------AAAUAAG-UUUAUGUUCGCAUGAUAUCAUAUC---UUAUACAAACCGGUCACGCUGGCAUCGCUAUCGCCCAUA--UUU-UUGGCAGUUGCCUGCGGACCACUUUC --------------..(((((-..((((.((....))...)))).)---)))).......((((.(((.((((..((((.........--...-.))))...)))).)))))))...... ( -26.42) >DroSec_CAF1 5459 99 - 1 --------------AUAUCAU-UUUGGAAUCUGAAGAUUU-UUUUC---UUAUACAAACCGGUCACGCUGGCAUCUCUAUCGCCCAUA--UUUUUUGGUAGUUGCCUGCGGACCACUUUC --------------.......-...((......((((...-...))---)).......))((((.(((.((((...((((((......--.....)))))).)))).)))))))...... ( -26.12) >DroAna_CAF1 7899 90 - 1 --------------AGAUAAC-CU-------------UUU-CUUUCUCAGUGUACAAACCGGUCACGCUACCACCGCUAUCGACAAUACUUUU-UUGGUAGUUGCCUUUGGGCCACUUUC --------------.......-..-------------...-.......(((((((......)).)))))......((((((((.((....)).-)))))))).(((....)))....... ( -14.20) >DroSim_CAF1 5688 98 - 1 --------------AUAUCAU-UUUGGAAUCUGAAGAUUU-GUUUC---UUAUACAAACCGGUCACGCUGGCAUCGCUAUCGCCCAUA--UUU-UUGGUAGUUGCCUGCGGACCACUUUC --------------.......-...((......((((...-...))---)).......))((((.(((.((((..(((((((......--...-.))))))))))).)))))))...... ( -26.92) >DroEre_CAF1 5601 113 - 1 AGCUGCUCUUCGUGAUAUCAUUUGUGGUAUCUGAAGAUUU-UGCUC---GUAUACAAACCGGUCACGCGGGCAUCGCUAUCGCCCAUA--UUU-UUGGUAGUUGCCUGCCGACCACUUUC (((...((((((.(((((((....)))))))))))))...-.))).---...........((((..(((((((..(((((((......--...-.)))))))))))))).))))...... ( -38.20) >DroYak_CAF1 6089 101 - 1 AGCUGCUCUU------------UUUGGCAUCUGAAAAUUU-UGUUU---GUAUACAAACCGGUCACGCUGGCAUCGCUAUCGCCCAUA--UUU-UUGGUAGUUGCCUGCGGACCACUUUC ((.((((...------------...)))).))........-.((((---(....))))).((((.(((.((((..(((((((......--...-.))))))))))).)))))))...... ( -30.30) >consensus ______________AUAUCAU_UUUGGAAUCUGAAGAUUU_UUUUC___GUAUACAAACCGGUCACGCUGGCAUCGCUAUCGCCCAUA__UUU_UUGGUAGUUGCCUGCGGACCACUUUC ............................................................((((.(((.((((..(((((((.............))))))))))).)))))))...... (-18.73 = -19.45 + 0.72)
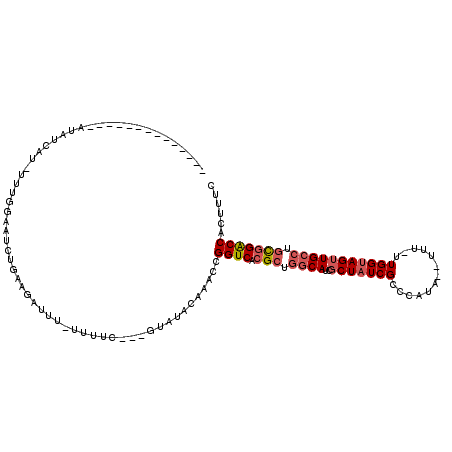
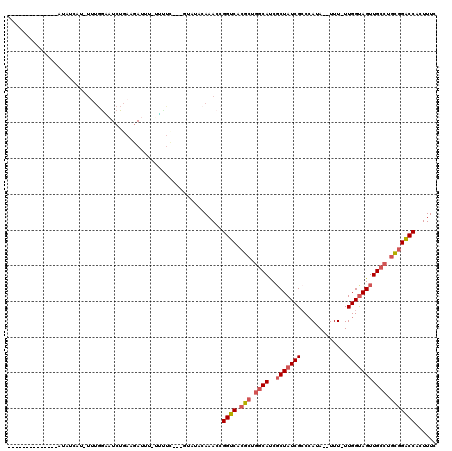
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:39 2006