| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,416,076 – 3,416,326 |
| Length | 250 |
| Max. P | 0.785758 |

| Location | 3,416,076 – 3,416,166 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.64 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -14.71 |
| Energy contribution | -13.74 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3416076 90 + 22407834 CAUA---------UCUCUGGU----CUUCAGCGGCUUUAU----------GUCUAUU--UUUGUAACAUUAUUUUCGAGACGAGCCUCGUUUAAGUGAU--GUUGGGU---CACUGGAGA ....---------((((..((----..((...(((.....----------)))....--....(((((((((((.((((......))))...)))))))--)))))).---.))..)))) ( -25.90) >DroSec_CAF1 5034 88 + 1 CAUA---------UCUCUGGU----CUUCAGGGGCUUUAU----------GCCUAUU--UUUGUAACAUUAUUUUCGAGACGAGCCUCGUUUAAGUGAU--GUUGGGU---CACUGGA-- ....---------..((..((----..((..((((.....----------))))...--....(((((((((((.((((......))))...)))))))--)))))).---.))..))-- ( -25.10) >DroAna_CAF1 7496 103 + 1 CAAAGUGGCAGGGCCUCUGGUCCGUCUUGUAGACUUUUAU----------GUUUAUUUCUUUGUGGCAUUAUUUUCGAGAUGAGCCUGGU-UAAGUGGU--GUUGGCU---CAUACGAA- ....((.((((((((...))))).....((((((......----------)))))).....))).))......((((..(((((((....-........--...))))---))).))))- ( -27.76) >DroSim_CAF1 5252 93 + 1 CAUA---------UCUCUGGU----CUUCAGGGGCUUUAU----------GCCUAUU--UUUGUAACAUUAUUUUCGAGACGAGCCUCGUUUAAGUGAU--GUUGGGUCGUCACUGGAGA ....---------.......(----((((((((((.....----------))))...--..(.(((((((((((.((((......))))...)))))))--)))).)......))))))) ( -28.40) >DroEre_CAF1 5192 89 + 1 CAUA---------UCUCUGGU----CUUCAGAGGCCUUAU----------GCCUAUU--UUUGUAACAUUAUUUUCGAGACGAGCCUCGC-UAAGUGAU--GUUGGGU---CACUGGAGA ....---------((((..((----..((..((((.....----------))))...--....(((((((((((.((((......)))).-.)))))))--)))))).---.))..)))) ( -28.50) >DroYak_CAF1 5639 102 + 1 CAUA---------UCUCUGCU----CUCGAGGGGCUUUAUGUUUUGAGGUGCCUAUU--UUUGUAACAUUAUUUUCGAGACGAGCCUCGUUUAAGUGAUGUGUUGGGU---CACUGGAGA ....---------((((((((----(...(((.((((((.....)))))).)))...--......(((((((((.((((......))))...)))))))))...))))---....))))) ( -29.00) >consensus CAUA_________UCUCUGGU____CUUCAGGGGCUUUAU__________GCCUAUU__UUUGUAACAUUAUUUUCGAGACGAGCCUCGUUUAAGUGAU__GUUGGGU___CACUGGAGA .........................((((((((((...............)))).........(((((((((((.((((......))))...)))))))..))))........)))))). (-14.71 = -13.74 + -0.96)
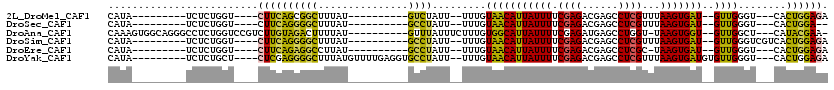
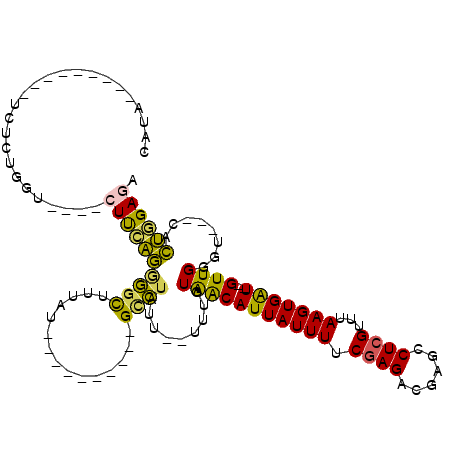
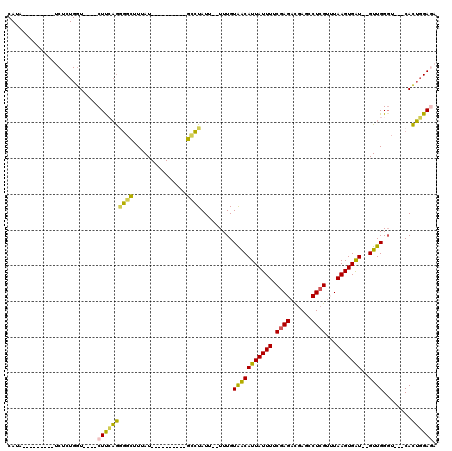
| Location | 3,416,166 – 3,416,286 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -23.63 |
| Energy contribution | -26.26 |
| Covariance contribution | 2.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3416166 120 + 22407834 UAUAUGCACAUGGGGCUUGCGUGUCAGUUGGUCAAGUGGACAGAGGUGAAACAUUUCGGUUUUAAACACUCUGUCUAAUGACUUGGGAUCAGCCAAGCCAAGAGACUCUGUGACCUACGA ......((((.((((((((.(((((....(((((..(((((((((.((((((......))))))....))))))))).)))))...)))..))))))))......)).))))........ ( -38.50) >DroSec_CAF1 5122 104 + 1 --------------GCUUGCGUGUCAGUUGGUCAAGUGGACAGAGGUGAAACAUUUCGGUUUUAAACACUCCGUCUAAUGGCUUGGGAUCACCCAAGCCAA--GUCUCUGUGACCUAAGA --------------...............(((((..(((((.(((.((((((......))))))....))).))))).(((((((((....))))))))).--.......)))))..... ( -36.50) >DroSim_CAF1 5345 110 + 1 UACAU--------GGCUUGCGUGUCAGUUGGUCAAGUGGACAGAGGUGAAACAUUUCGGUUUUAAACACUCCGUCUAAUGGCUUGGGAUCAGCCAAGCCAA--GACUCUGUGACCUAAGA ....(--------(((......))))...(((((..(((((.(((.((((((......))))))....))).))))).((((((((......)))))))).--.......)))))..... ( -37.00) >DroEre_CAF1 5281 112 + 1 UACC------UGACGCUUGCGUGUCAGUUGGUCAAGUGGCCAGAGGUGAAACAUUUCGGCUUUAAACAUUCUGCCUGCUGGCUUGGGAUCAGCCAAGCCAC--GACUUUGUGGCCCAUGA ...(------((((((....)))))))....(((.(.((((((((((..........(((............)))((.((((((((......)))))))))--)))))).)))))).))) ( -40.00) >DroYak_CAF1 5741 95 + 1 UACUU-----UGGGGUUUGCGUGUCAGUUGGUCAAGUGGCCAGAGGUGAAACAUUUCGGUUUUAAACAUACUG--------CUUGGGAUCAG------------ACUUGGUGACCUCUGA .....-----.((((((..((.(((...(((((....)))))((((((...))))))((((((((.(.....)--------.)))))))).)------------)).))..))))))... ( -25.00) >consensus UACAU_____UG_GGCUUGCGUGUCAGUUGGUCAAGUGGACAGAGGUGAAACAUUUCGGUUUUAAACACUCUGUCUAAUGGCUUGGGAUCAGCCAAGCCAA__GACUCUGUGACCUAAGA .............................(((((..(((((((((.((((((......))))))....))))))))).((((((((......))))))))..........)))))..... (-23.63 = -26.26 + 2.63)


| Location | 3,416,166 – 3,416,286 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -19.75 |
| Energy contribution | -21.78 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3416166 120 - 22407834 UCGUAGGUCACAGAGUCUCUUGGCUUGGCUGAUCCCAAGUCAUUAGACAGAGUGUUUAAAACCGAAAUGUUUCACCUCUGUCCACUUGACCAACUGACACGCAAGCCCCAUGUGCAUAUA ..((.(((((.((.......((((((((......))))))))...(((((((.((...((((......)))).)))))))))..))))))).))......(((.........)))..... ( -31.60) >DroSec_CAF1 5122 104 - 1 UCUUAGGUCACAGAGAC--UUGGCUUGGGUGAUCCCAAGCCAUUAGACGGAGUGUUUAAAACCGAAAUGUUUCACCUCUGUCCACUUGACCAACUGACACGCAAGC-------------- .....(((((.((....--.(((((((((....)))))))))...(((((((.((...((((......)))).)))))))))..)))))))........(....).-------------- ( -35.90) >DroSim_CAF1 5345 110 - 1 UCUUAGGUCACAGAGUC--UUGGCUUGGCUGAUCCCAAGCCAUUAGACGGAGUGUUUAAAACCGAAAUGUUUCACCUCUGUCCACUUGACCAACUGACACGCAAGCC--------AUGUA .....(((((.((....--.((((((((......))))))))...(((((((.((...((((......)))).)))))))))..))))))).....((((....)..--------.))). ( -32.30) >DroEre_CAF1 5281 112 - 1 UCAUGGGCCACAAAGUC--GUGGCUUGGCUGAUCCCAAGCCAGCAGGCAGAAUGUUUAAAGCCGAAAUGUUUCACCUCUGGCCACUUGACCAACUGACACGCAAGCGUCA------GGUA (((..(((((.......--.((((((((......))))))))(.(((..((((((((.......))))))))..)))))))))...)))....(((((.(....).))))------)... ( -39.70) >DroYak_CAF1 5741 95 - 1 UCAGAGGUCACCAAGU------------CUGAUCCCAAG--------CAGUAUGUUUAAAACCGAAAUGUUUCACCUCUGGCCACUUGACCAACUGACACGCAAACCCCA-----AAGUA ((((.(((((....((------------(.((....(((--------((...((........))...)))))....)).)))....)))))..)))).............-----..... ( -14.00) >consensus UCAUAGGUCACAGAGUC__UUGGCUUGGCUGAUCCCAAGCCAUUAGACAGAGUGUUUAAAACCGAAAUGUUUCACCUCUGUCCACUUGACCAACUGACACGCAAGCC_CA_____AUGUA .....(((((..........((((((((......))))))))...(((((((.((...((((......)))).)))))))))....)))))............................. (-19.75 = -21.78 + 2.03)


| Location | 3,416,206 – 3,416,326 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.96 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -16.56 |
| Energy contribution | -18.60 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3416206 120 - 22407834 CAAAUUCUGAAAUCUAAUUUAGAUUCUUAAGUCCUAGGAGUCGUAGGUCACAGAGUCUCUUGGCUUGGCUGAUCCCAAGUCAUUAGACAGAGUGUUUAAAACCGAAAUGUUUCACCUCUG .....((((..(((((.....((((((((.....)))))))).)))))..))))((((..((((((((......))))))))..))))((((.((...((((......)))).)))))). ( -36.00) >DroSec_CAF1 5148 117 - 1 -AAAUUCUGAAAUCUAAUUUAGGUUCUUAAGCCCUAGGAGUCUUAGGUCACAGAGAC--UUGGCUUGGGUGAUCCCAAGCCAUUAGACGGAGUGUUUAAAACCGAAAUGUUUCACCUCUG -......((((((...((((.((((.((((((.((....((((.(((((.....)))--))((((((((....))))))))...))))..)).)))))))))).))))))))))...... ( -36.70) >DroSim_CAF1 5377 117 - 1 -AAAUUCUGAAAUCUAAUUUAGGUCCUUAAGCCCUAGGAGUCUUAGGUCACAGAGUC--UUGGCUUGGCUGAUCCCAAGCCAUUAGACGGAGUGUUUAAAACCGAAAUGUUUCACCUCUG -....((((..((((((....(.((((........)))).).))))))..))))(((--(((((((((......))))))))..))))((((.((...((((......)))).)))))). ( -32.70) >DroEre_CAF1 5315 117 - 1 -AAAUUCAACCAUCAAACUUGGUGUCUUAAGCACUAGGAGUCAUGGGCCACAAAGUC--GUGGCUUGGCUGAUCCCAAGCCAGCAGGCAGAAUGUUUAAAGCCGAAAUGUUUCACCUCUG -........((((.(..((((((((.....))))))))..).))))(((((......--)))))..((((.......))))((.(((..((((((((.......))))))))..))))). ( -33.60) >DroYak_CAF1 5776 99 - 1 -AAAAUCAGCCAUCUAACUUGGGCUCUUAAGCCCUAGGAGUCAGAGGUCACCAAGU------------CUGAUCCCAAG--------CAGUAUGUUUAAAACCGAAAUGUUUCACCUCUG -................((.(((((....))))).))....(((((((......(.------------.((....))..--------)...............(((....)))))))))) ( -20.20) >consensus _AAAUUCUGAAAUCUAAUUUAGGUUCUUAAGCCCUAGGAGUCAUAGGUCACAGAGUC__UUGGCUUGGCUGAUCCCAAGCCAUUAGACAGAGUGUUUAAAACCGAAAUGUUUCACCUCUG ....(((..............((((((((.....))))))))...((((((...(((...((((((((......))))))))...)))...)))......)))))).............. (-16.56 = -18.60 + 2.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:37 2006