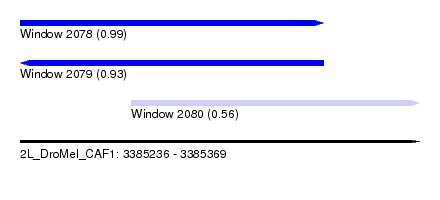
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,385,236 – 3,385,369 |
| Length | 133 |
| Max. P | 0.988832 |

| Location | 3,385,236 – 3,385,337 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -9.85 |
| Energy contribution | -9.49 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3385236 101 + 22407834 AGUGAGU--GCUGCAAGUCGAAUGGAAUUCG-AUUCGAUUUGAUUUCGUAUUUAUAUUCAAUUUAUUCUAGAUAUUUGCUGCUCUAUGCAGCAUUUUCCACUCC .((((((--((..((((((((((.(....).-)))))))))).....))))))))...............((....((((((.....))))))...))...... ( -29.00) >DroPse_CAF1 13343 90 + 1 ------U--UCUAAAAACUCGAUCAAAC------UCGAUUUGAUUUCGUAUUUAUAUUCAAUUUCUCUUAGAUAUUUGCUGUUCUAUGCAGCAUUUUCUACAUC ------.--..((((.((..(((((((.------....)))))))..)).))))..............((((....((((((.....))))))...)))).... ( -15.60) >DroEre_CAF1 8052 98 + 1 AGUGAGUAGACUGCAAGUCGAAUAGAAUUC------GAUUUGAUUUCGUAUUUAUAUUCAAUUUAUUCUAGAUAUUUGCUGCUCUAUGCAGCAUUUUCCACUCC .(((((((((...((((((((((...))))------))))))...)).)))))))...............((....((((((.....))))))...))...... ( -24.80) >DroYak_CAF1 8185 96 + 1 AGUGAGU--GCUGCAAGUCGAAUAGAAUUC------GAUUUGAUUUCGUAUUUAUAUUCAAUUUAUUCUAGAUAUUUGCUGCUCUAUGCAGCAUUUUCCACUCC .((((((--((..((((((((((...))))------)))))).....))))))))...............((....((((((.....))))))...))...... ( -25.30) >DroAna_CAF1 13686 95 + 1 CCCGAGU--GGAGCA-------UCGAAUUCGAAUUCGAUUUGAUUUCGUAUUUAUAUUCAAUUUAUUCUAGAUAUUUGCUGCUCCACGCAGCGUUUUCCACUCC ...((((--((((..-------..((((..((((.(((.......))).))))..))))..................(((((.....)))))...)))))))). ( -27.00) >DroPer_CAF1 12770 90 + 1 ------U--UCUAAAAACUCGAUCAAAC------UCGAUUUGAUUUCGUAUUUAUAUUCAAUUUCUCUUAGAUAUUUGCUGUUCUAUGCAGCAUUUUCUACAUC ------.--..((((.((..(((((((.------....)))))))..)).))))..............((((....((((((.....))))))...)))).... ( -15.60) >consensus AGUGAGU__GCUGCAAGUCGAAUCGAAUUC____UCGAUUUGAUUUCGUAUUUAUAUUCAAUUUAUUCUAGAUAUUUGCUGCUCUAUGCAGCAUUUUCCACUCC ...............................................(((((((..............))))))).((((((.....))))))........... ( -9.85 = -9.49 + -0.36)


| Location | 3,385,236 – 3,385,337 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -8.82 |
| Energy contribution | -9.10 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3385236 101 - 22407834 GGAGUGGAAAAUGCUGCAUAGAGCAGCAAAUAUCUAGAAUAAAUUGAAUAUAAAUACGAAAUCAAAUCGAAU-CGAAUUCCAUUCGACUUGCAGC--ACUCACU .(((((.....((((((.....))))))..................................(((.((((((-.(....).)))))).)))...)--))))... ( -23.40) >DroPse_CAF1 13343 90 - 1 GAUGUAGAAAAUGCUGCAUAGAACAGCAAAUAUCUAAGAGAAAUUGAAUAUAAAUACGAAAUCAAAUCGA------GUUUGAUCGAGUUUUUAGA--A------ ...........(((((.......)))))....((((((((...((((...(((((.(((.......))).------))))).)))).))))))))--.------ ( -16.70) >DroEre_CAF1 8052 98 - 1 GGAGUGGAAAAUGCUGCAUAGAGCAGCAAAUAUCUAGAAUAAAUUGAAUAUAAAUACGAAAUCAAAUC------GAAUUCUAUUCGACUUGCAGUCUACUCACU .(((((((...((((((.....))))))...........(((.(((((((..(((.(((.......))------).))).))))))).)))...)))))))... ( -24.40) >DroYak_CAF1 8185 96 - 1 GGAGUGGAAAAUGCUGCAUAGAGCAGCAAAUAUCUAGAAUAAAUUGAAUAUAAAUACGAAAUCAAAUC------GAAUUCUAUUCGACUUGCAGC--ACUCACU .(((((.....((((((.....))))))...........(((.(((((((..(((.(((.......))------).))).))))))).)))...)--))))... ( -22.00) >DroAna_CAF1 13686 95 - 1 GGAGUGGAAAACGCUGCGUGGAGCAGCAAAUAUCUAGAAUAAAUUGAAUAUAAAUACGAAAUCAAAUCGAAUUCGAAUUCGA-------UGCUCC--ACUCGGG .(((((((....(((((.....)))))......................................(((((((....))))))-------)..)))--))))... ( -27.10) >DroPer_CAF1 12770 90 - 1 GAUGUAGAAAAUGCUGCAUAGAACAGCAAAUAUCUAAGAGAAAUUGAAUAUAAAUACGAAAUCAAAUCGA------GUUUGAUCGAGUUUUUAGA--A------ ...........(((((.......)))))....((((((((...((((...(((((.(((.......))).------))))).)))).))))))))--.------ ( -16.70) >consensus GGAGUGGAAAAUGCUGCAUAGAGCAGCAAAUAUCUAGAAUAAAUUGAAUAUAAAUACGAAAUCAAAUCGA____GAAUUCGAUUCGACUUGCAGC__ACUCACU ....((((...((((((.....))))))....)))).................................................................... ( -8.82 = -9.10 + 0.28)


| Location | 3,385,273 – 3,385,369 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -16.29 |
| Consensus MFE | -9.85 |
| Energy contribution | -9.49 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3385273 96 + 22407834 UGAUUUCGUAUUUAUAUUCAAUUUAUUCUAGAUAUUUGCUGCUCUAUGCAGCAUUUUCCACUCCGAUAU--GCUUUCCUUUGGCUGGCGCUUGCCUUG .((...((((((........(((((...)))))...((((((.....))))))...........)))))--)...))....(((........)))... ( -15.70) >DroPse_CAF1 13369 93 + 1 UGAUUUCGUAUUUAUAUUCAAUUUCUCUUAGAUAUUUGCUGUUCUAUGCAGCAUUUUCUACAUCGAAAUCUACUUCCACUU----CG-AUUUGCUUUG .(((((((....................((((....((((((.....))))))...))))...)))))))...........----..-.......... ( -15.40) >DroSec_CAF1 8082 94 + 1 UGAUUUCGUAUUUAUAUUCAAUUUAUUCUAGAUAUUUGCUGCUCUAUGCAGCAUUUUCCACUCCGAUAU--GCUUUCCU-UGGCUCG-GCUUGGCUUG .......(((((((..............))))))).((((((.....))))))....(((..((((...--(((.....-.))))))-)..))).... ( -19.74) >DroEre_CAF1 8086 95 + 1 UGAUUUCGUAUUUAUAUUCAAUUUAUUCUAGAUAUUUGCUGCUCUAUGCAGCAUUUUCCACUCCGAUAU--GCUUUCCUUUGGCUCA-GCUUGCCUUG .((...((((((........(((((...)))))...((((((.....))))))...........)))))--)...))....(((...-....)))... ( -15.40) >DroAna_CAF1 13717 91 + 1 UGAUUUCGUAUUUAUAUUCAAUUUAUUCUAGAUAUUUGCUGCUCCACGCAGCGUUUUCCACUCCGUGUU--GCUCCACUACAGCUC-----UGAGUUG ..............................((....((((((.....))))))...)).((((.(.(((--(........)))).)-----.)))).. ( -16.10) >DroPer_CAF1 12796 93 + 1 UGAUUUCGUAUUUAUAUUCAAUUUCUCUUAGAUAUUUGCUGUUCUAUGCAGCAUUUUCUACAUCGAAAUCUACUUCCACUU----CG-AUUUGCUUUG .(((((((....................((((....((((((.....))))))...))))...)))))))...........----..-.......... ( -15.40) >consensus UGAUUUCGUAUUUAUAUUCAAUUUAUUCUAGAUAUUUGCUGCUCUAUGCAGCAUUUUCCACUCCGAUAU__GCUUCCCUUUGGCUCG_GCUUGCCUUG .......(((((((..............))))))).((((((.....))))))............................................. ( -9.85 = -9.49 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:18 2006