
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,379,886 – 3,379,989 |
| Length | 103 |
| Max. P | 0.563783 |

| Location | 3,379,886 – 3,379,989 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.45 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -10.73 |
| Energy contribution | -11.06 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3379886 103 + 22407834 AAAAGGAGUCGGCCAAGUGGCUCUCAAGCUGCAGCCACCACUGCAGCAUAACAGGCACAAAGUUGUCGCAAAAACUUG---------GCCA-------GAAAACGCAGAUGGAGCAACU- ....(...((((((((((.((......(((((((......)))))))......((((......))))))....)))))---------))).-------))...)((.......))....- ( -34.80) >DroSec_CAF1 2729 107 + 1 AAAAGGAGUCGGCCAAGUGGCUCUCAAACUG------------CAGCAUAACAGGCACAAAGUUGUCGCAAAAACUUGGAUCCGCUGGCCAGGAAACUGAAAACGCUGAUGGAGCAACU- ....(...(((((((.((((.((.(((.(((------------........)))(((((....))).))......))))).)))))))))((....))))...)(((.....)))....- ( -29.00) >DroSim_CAF1 2749 119 + 1 AAAAGGAGUCGGCCAAGUGGCUCUCAAACUGCAGCCACCACUGCAGCAUAACAGGCACAAAGUUGUCGCAAAAACUUGGAUCCGCUGGCCAGGAAACUGAAAACGCUGAUGGAGCAACU- ....(...(((((((.((((.((.(((.((((((......)))))).......((((......))))........))))).)))))))))((....))))...)(((.....)))....- ( -36.30) >DroEre_CAF1 2632 115 + 1 AAAAGGAGUCG-CAAAGUGGCUCUCAAACUGCAGCCUGCACUGC---AUAGCAGGCACAAAGUUGCCGCAAAAACUUGGAGCCGCUCGCCAGGAAACAGAAAACGCUGAUGGAGCAACU- ......(((.(-(..(((((((((.........((((((.....---...))))))...(((((........))))))))))))))..(((.....(((......))).))).)).)))- ( -35.90) >DroYak_CAF1 2763 116 + 1 AAAAGGAGUCGGCAAAGUGGCUCUCAAACUGCAGCCAACAAUGC---AGAGCAGGCACAAAGUUGUCGCAAAAACUUGGAGCCGCUCGCAAGGAAACUAAAAACGCUGAUGGAGCAACU- .......((((((..((((((((.(((.(((((........)))---)).((.((((......))))))......)))))))))))....((....))......)))))).........- ( -38.00) >DroAna_CAF1 2803 92 + 1 GGAAAGAGCCUGCGAAGGGGCUCUCAAACUGGAGUCAAUAC-A---CAUAACUGGCACAAAGUUGACGUGGAAAGUUCGAG------------------------GAGUUGCAGCAACUU ....(((((((......)))))))....((.(((.(....(-(---(.(((((.......)))))..)))....)))).))------------------------((((((...)))))) ( -21.40) >consensus AAAAGGAGUCGGCCAAGUGGCUCUCAAACUGCAGCCAACACUGC__CAUAACAGGCACAAAGUUGUCGCAAAAACUUGGAGCCGCU_GCCAGGAAACUGAAAACGCUGAUGGAGCAACU_ ....(((((((......))))))).....(((.(((.................)))...(((((........)))))....................................))).... (-10.73 = -11.06 + 0.34)

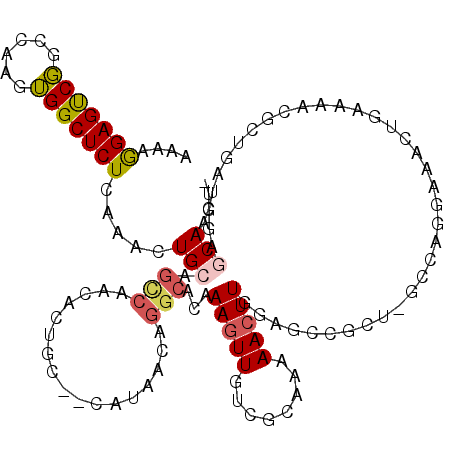

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:13 2006