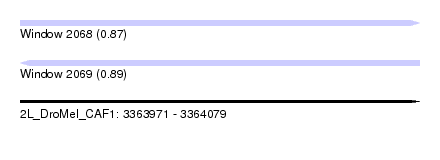
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,363,971 – 3,364,079 |
| Length | 108 |
| Max. P | 0.888869 |

| Location | 3,363,971 – 3,364,079 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -42.74 |
| Consensus MFE | -27.80 |
| Energy contribution | -29.05 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
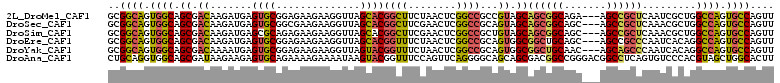
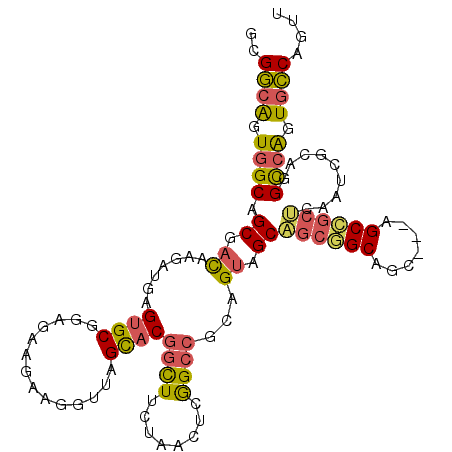
>2L_DroMel_CAF1 3363971 108 + 22407834 AACUGGCACUGGCCAGCGAUUGAGCGGCU---UCUGCCGCUGCUACGGCGGCCGAGUUAGAAGCCGUGCUAACCUUCUUCUCCGCACUCAUCUUGUCGCUGCCACUGCCGC ....((((.((((.((((((.((..((((---(((((((((((....)))).)).).))))))))((((..............))))...))..)))))))))).)))).. ( -50.74) >DroSec_CAF1 54694 108 + 1 AACUGGCACUGGCCAGCGUUUGAGCGGCU---GCUGCCGCUGCUACUGCGGCCGAGUUCGAAGCCGUGCUAACCUUCUUCGCCGCACUCAUCUUGUCGCUGCCACUGCCGC ....((((.((((.((((....((((((.---...)))))).....((((((.(((.....(((...)))......))).))))))..........)))))))).)))).. ( -45.20) >DroSim_CAF1 50748 108 + 1 AACUGGCACUGGCCAGCGUUUGAGCGGCU---GCUGCCGCUGCUACAGCGGCCGAGUUCGAAGCCGUGCUAACCUUCUUCUCUGCGCUCAUCUUGUCGCUGCCACUGCCGC ....((((.((((.((((..(((((((((---((((.........))))))))(((...((((..((....))))))..)))...)))))......)))))))).)))).. ( -47.10) >DroEre_CAF1 49155 108 + 1 AACUGGCACUGGCCUGUGAUUGGGCGGCU---GCUGCAGCCGCCACUGCGGCCGAGUUAGAAACCGUGCUAACCUUCUUCUCCGCACUCAUCUUGUCGCUGCCACUGCCGC ....((((.((((..(((((..(((((((---(...))))))))..(((((..(((((((........)))))..))....)))))........))))).)))).)))).. ( -44.80) >DroYak_CAF1 50395 108 + 1 AACUGGCACUGGCCUGUGAUUGGGCUGCU---GUUGCAGCCGCCACUGCGGCCGAGUUAGAAACCGUACUAACCUUCUUCUCCGCACUCAUUUUGUCGCUGCCACUGCCGC ....((((.((((..(((((..((((((.---...)))))).....(((((..(((((((........)))))..))....)))))........))))).)))).)))).. ( -41.00) >DroAna_CAF1 58832 111 + 1 AAGUGCCAGCUACGUGGGACACUGAGGCCGUCCCGGCCGUCGCUGCUGCCCCUGAACUGGAAACCGUACUUAUUUUCUUUUCUGCACUCUUCUUAUCGCUGCCACCUGCAG ..(((.((((..((.(((.((.((((((((...))))).))).))...))).)).((.(....).))..............................)))).)))...... ( -27.60) >consensus AACUGGCACUGGCCAGCGAUUGAGCGGCU___GCUGCCGCCGCUACUGCGGCCGAGUUAGAAACCGUGCUAACCUUCUUCUCCGCACUCAUCUUGUCGCUGCCACUGCCGC ...(((((.((((..(((((..((((((.......))))))......((((..(((((((........)))))..))....)))).........))))).)))).))))). (-27.80 = -29.05 + 1.25)

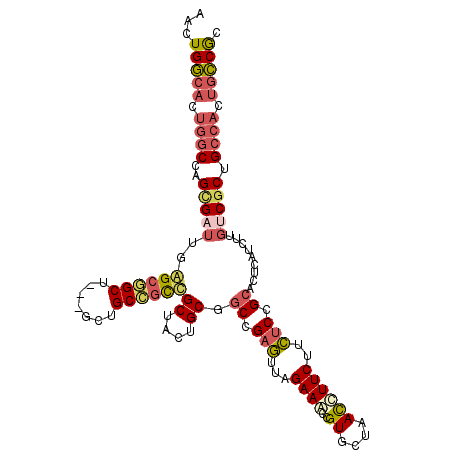
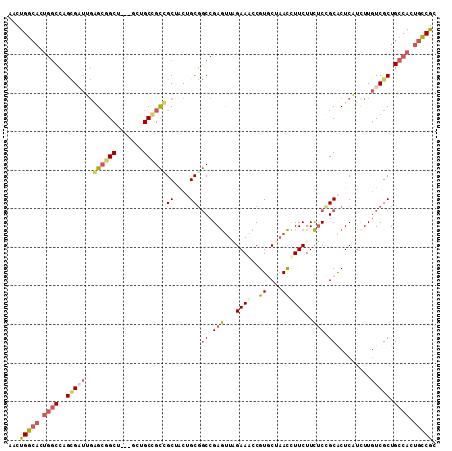
| Location | 3,363,971 – 3,364,079 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -45.43 |
| Consensus MFE | -33.47 |
| Energy contribution | -33.14 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3363971 108 - 22407834 GCGGCAGUGGCAGCGACAAGAUGAGUGCGGAGAAGAAGGUUAGCACGGCUUCUAACUCGGCCGCCGUAGCAGCGGCAGA---AGCCGCUCAAUCGCUGGCCAGUGCCAGUU ..((((.(((((((((........((((..............))))(((((((......(((((.......))))))))---))))......))))).)))).)))).... ( -48.14) >DroSec_CAF1 54694 108 - 1 GCGGCAGUGGCAGCGACAAGAUGAGUGCGGCGAAGAAGGUUAGCACGGCUUCGAACUCGGCCGCAGUAGCAGCGGCAGC---AGCCGCUCAAACGCUGGCCAGUGCCAGUU ..((((.((((((((........(.((((((((((..(.......)..))))(....).)))))).).(.((((((...---.)))))))...)))).)))).)))).... ( -49.80) >DroSim_CAF1 50748 108 - 1 GCGGCAGUGGCAGCGACAAGAUGAGCGCAGAGAAGAAGGUUAGCACGGCUUCGAACUCGGCCGCUGUAGCAGCGGCAGC---AGCCGCUCAAACGCUGGCCAGUGCCAGUU ..((((.((((((((......(((((((.(((..((((.(......).))))...))).(((((((...)))))))...---.).))))))..)))).)))).)))).... ( -49.50) >DroEre_CAF1 49155 108 - 1 GCGGCAGUGGCAGCGACAAGAUGAGUGCGGAGAAGAAGGUUAGCACGGUUUCUAACUCGGCCGCAGUGGCGGCUGCAGC---AGCCGCCCAAUCACAGGCCAGUGCCAGUU ..((((.((((.(((......(((((....(((((..(.......)..))))).)))))..))).(.((((((((...)---))))))))........)))).)))).... ( -44.90) >DroYak_CAF1 50395 108 - 1 GCGGCAGUGGCAGCGACAAAAUGAGUGCGGAGAAGAAGGUUAGUACGGUUUCUAACUCGGCCGCAGUGGCGGCUGCAAC---AGCAGCCCAAUCACAGGCCAGUGCCAGUU ..((((.((((..............(((((....((..(((((........)))))))..)))))((((.((((((...---.))))))...))))..)))).)))).... ( -44.00) >DroAna_CAF1 58832 111 - 1 CUGCAGGUGGCAGCGAUAAGAAGAGUGCAGAAAAGAAAAUAAGUACGGUUUCCAGUUCAGGGGCAGCAGCGACGGCCGGGACGGCCUCAGUGUCCCACGUAGCUGGCACUU ....(((((.((((((...((((.((((..............))))..))))....)).((((((.(...((.(((((...))))))).))))))).....)))).))))) ( -36.24) >consensus GCGGCAGUGGCAGCGACAAGAUGAGUGCGGAGAAGAAGGUUAGCACGGCUUCUAACUCGGCCGCAGUAGCAGCGGCAGC___AGCCGCUCAAUCGCAGGCCAGUGCCAGUU ..((((.((((.((.((.......((((..............))))((((........))))...)).))((((((.......)))))).........)))).)))).... (-33.47 = -33.14 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:07 2006