
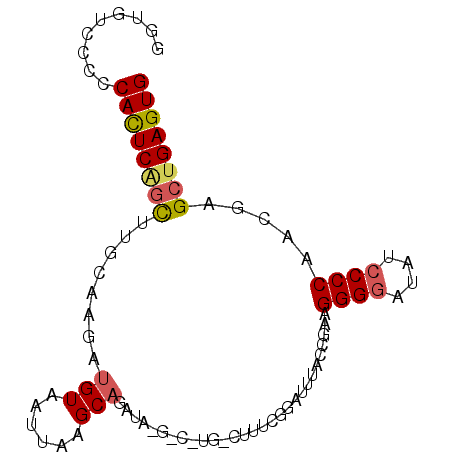
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,351,776 – 3,351,866 |
| Length | 90 |
| Max. P | 0.967270 |

| Location | 3,351,776 – 3,351,866 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 72.33 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -17.22 |
| Energy contribution | -17.54 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
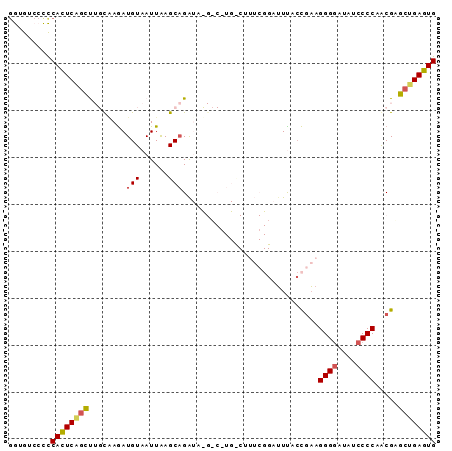
>2L_DroMel_CAF1 3351776 90 + 22407834 GGUGUCCCCCACUCGGCUUGUGAGAUGUAAUUAAGCAGAUACGGC-UGCCUUUCGGAUUUGCCGAAGGGGAUAUCCCCAACGUGCUGAGUG .........((((((((.(((.............((((......)-)))..(((((.....)))))((((....)))).))).)))))))) ( -32.00) >DroSec_CAF1 42591 91 + 1 GGUGUCCCCCACUCAGCUUGCAAGAUGUAAUUAAGCAGAUAUGUCGUGUCUUUCGGAUUCACCGAAGGGGAUAUCCCCAACGAGCUGAGUG .........(((((((((((................((((((...))))))(((((.....)))))((((....))))..))))))))))) ( -32.70) >DroSim_CAF1 38547 91 + 1 GGUGUCCCCCACUCAGCCUGCAAGAUGUAAUUAAGCAGAUAUGUCGUGUCUUUCGGAUUUACCGAAGGGGAUAUCCCCAACGAGCUGAGUG .........((((((((((((.............)))).....((((....(((((.....)))))((((....)))).)))))))))))) ( -33.52) >DroEre_CAF1 37277 70 + 1 GAUGUCCUUCACUCGGCC--CAAAAUGUAAUUAAGCAGA-----------------UGUU--UUUAGGGGAUAUCCCCAGCGAGCUGAGUG .........((((((((.--(....(((......)))..-----------------....--....((((....))))...).)))))))) ( -21.50) >DroYak_CAF1 38202 72 + 1 CAUGUCCUUCAUUCUUUU--UCAGAUGUAAUUGAGCUGG-----------------AUUCACCGAAGGGUAUAUCCCCAACGAGCUGAGUG ..................--.........(((.((((((-----------------((..(((....)))..))))......)))).))). ( -14.50) >consensus GGUGUCCCCCACUCAGCUUGCAAGAUGUAAUUAAGCAGAUA_G_C_UG_CUUUCGGAUUUACCGAAGGGGAUAUCCCCAACGAGCUGAGUG .........((((((((........(((......))).............................((((....)))).....)))))))) (-17.22 = -17.54 + 0.32)

| Location | 3,351,776 – 3,351,866 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 72.33 |
| Mean single sequence MFE | -25.99 |
| Consensus MFE | -12.61 |
| Energy contribution | -15.30 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
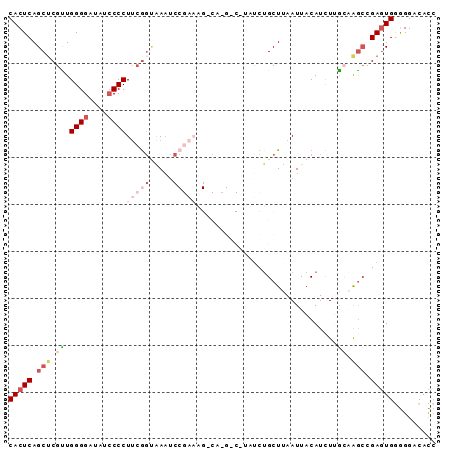
>2L_DroMel_CAF1 3351776 90 - 22407834 CACUCAGCACGUUGGGGAUAUCCCCUUCGGCAAAUCCGAAAGGCA-GCCGUAUCUGCUUAAUUACAUCUCACAAGCCGAGUGGGGGACACC (((((.((.....((((....))))(((((.....)))))(((((-(......))))))...............)).)))))(....)... ( -29.30) >DroSec_CAF1 42591 91 - 1 CACUCAGCUCGUUGGGGAUAUCCCCUUCGGUGAAUCCGAAAGACACGACAUAUCUGCUUAAUUACAUCUUGCAAGCUGAGUGGGGGACACC ((((((((((((.((((....))))(((((.....)))))....))))......(((.............))).))))))))(....)... ( -32.42) >DroSim_CAF1 38547 91 - 1 CACUCAGCUCGUUGGGGAUAUCCCCUUCGGUAAAUCCGAAAGACACGACAUAUCUGCUUAAUUACAUCUUGCAGGCUGAGUGGGGGACACC ((((((((((((.((((....))))(((((.....)))))....)))).....((((.............))))))))))))(....)... ( -35.32) >DroEre_CAF1 37277 70 - 1 CACUCAGCUCGCUGGGGAUAUCCCCUAAA--AACA-----------------UCUGCUUAAUUACAUUUUG--GGCCGAGUGAAGGACAUC ..((..(((((..((((....))))....--....-----------------...((((((.......)))--))))))))..))...... ( -19.60) >DroYak_CAF1 38202 72 - 1 CACUCAGCUCGUUGGGGAUAUACCCUUCGGUGAAU-----------------CCAGCUCAAUUACAUCUGA--AAAAGAAUGAAGGACAUG ..(((((....))))).......(((((((.....-----------------))............(((..--...)))..)))))..... ( -13.30) >consensus CACUCAGCUCGUUGGGGAUAUCCCCUUCGGUAAAUCCGAAAG_CA_G_C_UAUCUGCUUAAUUACAUCUUGCAAGCCGAGUGGGGGACACC (((((.(((.((.((((....))))(((((.....)))))..............................)).))).)))))......... (-12.61 = -15.30 + 2.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:05 2006