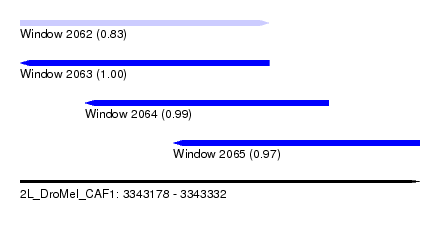
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,343,178 – 3,343,332 |
| Length | 154 |
| Max. P | 0.995108 |

| Location | 3,343,178 – 3,343,274 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 71.62 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.85 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
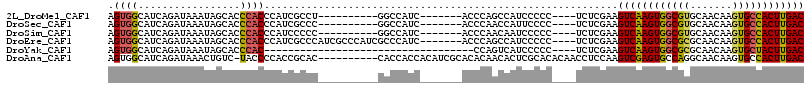
>2L_DroMel_CAF1 3343178 96 + 22407834 GUGG---UGGAGG--AAGGGAUGGCUUAUGCCAAACAGAGGCAGACGAUGUCGAGUCGAGACCAGUCAAGUGGCACUUGUUGCACGCCACUUGACUUCGAG .(((---(.....--...(..((((....))))..)...(((.(((...)))..)))...))))((((((((((...........))))))))))...... ( -31.70) >DroSec_CAF1 34003 101 + 1 GAGGUGGUGGGGGGAGCAGGGUGGCUUAUGCCAAGGAGGGGCAGACGAUGUCGAGUUGAGUCGAGUCAAGUGGCACUUGUUGCACGCCACUUGACUUCGAG ((((((((((.((.((((((((.((((.((((.......))))...(((.((((......))))))))))).)).)))))).).).)))))..)))))... ( -38.30) >DroSim_CAF1 30088 99 + 1 GAGGUGGUGGGGG--GCAGGGUGGCUUAUGCCAAAGAGGGGCAGACGAUGUCGAGUCGAGUCGAGUCAAGUGGCACUUGUUGCACGCCACUUGACUUCGAG .((((((((.(.(--(((((((.((((.((((.......))))...(((.((((......))))))))))).)).)))))).).))))))))......... ( -36.50) >DroEre_CAF1 28492 82 + 1 AAU---A----------GGGGGGUCUUAUGCCAAAGAGG-GUAUGCGAAGUCG-----CGUCGAGUCAAGUGGCACUUGUUGCGCGCCACUUGACUUCGAG ...---.----------(.((..((.((((((......)-))))).))..)).-----).((((((((((((((.(.......).)))))))))).)))). ( -33.10) >DroYak_CAF1 29367 85 + 1 GAGGUGA----------GAGGGGGUUUAUGCCAAAGAGG-GUAGACGAAGUCG-----AGUCGAGUCAAGUAGCACUUGUUGCGCGCCACUUGACUUCGAG .......----------......((((((.((.....))-))))))(((((((-----(((.(.((...(((((....)))))..)))))))))))))... ( -25.60) >DroAna_CAF1 36257 74 + 1 GAGG------A--------GGGAGCUGAGGUGAA--------CAACGAAGCCA-----AGUCGUGUCAAGUGGCACUUGUUGCCUGGCACUCGACUUGGAG ..(.------.--------(...((....))...--------)..)....(((-----((((((((((.(..((....))..).))))))..))))))).. ( -23.80) >consensus GAGG_____________GGGGGGGCUUAUGCCAAAGAGG_GCAGACGAAGUCG_____AGUCGAGUCAAGUGGCACUUGUUGCACGCCACUUGACUUCGAG ......................(((....)))............................((((((((((((((...........)))))))))).)))). (-17.60 = -18.85 + 1.25)

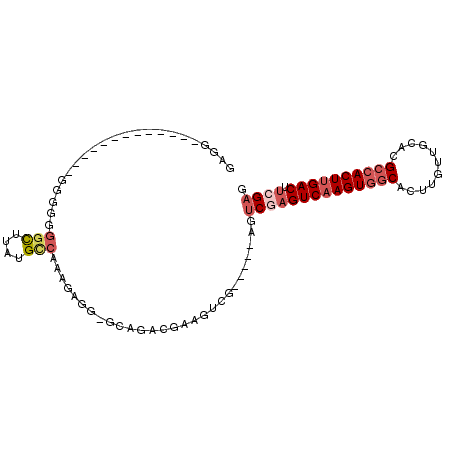
| Location | 3,343,178 – 3,343,274 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 71.62 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -18.94 |
| Energy contribution | -19.42 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3343178 96 - 22407834 CUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUGGUCUCGACUCGACAUCGUCUGCCUCUGUUUGGCAUAAGCCAUCCCUU--CCUCCA---CCAC .....((((((((((((.........))))))))))))(((...(((........))).......(..((((....))))..)...--.....)---)).. ( -28.80) >DroSec_CAF1 34003 101 - 1 CUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACUCAACUCGACAUCGUCUGCCCCUCCUUGGCAUAAGCCACCCUGCUCCCCCCACCACCUC .((((.(((((((((((.........)))))))))))))))........(((...)))..........((((....))))..................... ( -27.00) >DroSim_CAF1 30088 99 - 1 CUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACUCGACUCGACAUCGUCUGCCCCUCUUUGGCAUAAGCCACCCUGC--CCCCCACCACCUC .((((.(((((((((((.........)))))))))))))))...(((........)))..........((((....))))......--............. ( -28.50) >DroEre_CAF1 28492 82 - 1 CUCGAAGUCAAGUGGCGCGCAACAAGUGCCACUUGACUCGACG-----CGACUUCGCAUAC-CCUCUUUGGCAUAAGACCCCCC----------U---AUU .((((.((((((((((((.......)))))))))))))))).(-----((....)))....-..((((......))))......----------.---... ( -31.70) >DroYak_CAF1 29367 85 - 1 CUCGAAGUCAAGUGGCGCGCAACAAGUGCUACUUGACUCGACU-----CGACUUCGUCUAC-CCUCUUUGGCAUAAACCCCCUC----------UCACCUC .((((.((((((((((((.......))))))))))))))))..-----.......(((...-.......)))............----------....... ( -25.70) >DroAna_CAF1 36257 74 - 1 CUCCAAGUCGAGUGCCAGGCAACAAGUGCCACUUGACACGACU-----UGGCUUCGUUG--------UUCACCUCAGCUCCC--------U------CCUC ..((((((((.((....((((.....)))).....)).)))))-----)))....((((--------.......))))....--------.------.... ( -19.90) >consensus CUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACU_____CGACAUCGUCUGC_CCUCUUUGGCAUAAGCCACCCC_____________CCUC .((((.((((((((((((.......))))))))))))))))..............(((...........)))............................. (-18.94 = -19.42 + 0.47)

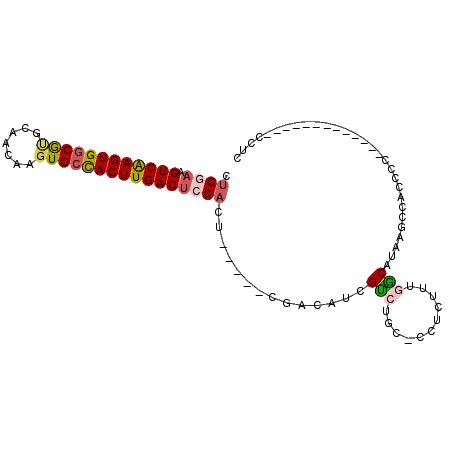
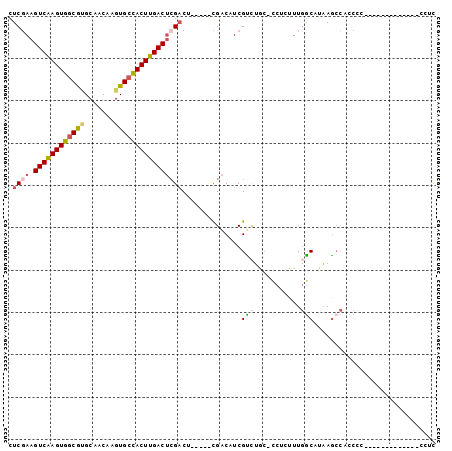
| Location | 3,343,203 – 3,343,297 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 72.49 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3343203 94 - 22407834 -----GGCCAUC-------ACCCAGCCAUCCCCC----UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUGGUCUCGACUCGACAUCGUCUGCCUCUGUUUGG -----(((....-------.....))).....((----...((.((((((((((((.........))))))))))))(((...(((........))).)))..))...)) ( -25.90) >DroSec_CAF1 34033 94 - 1 -----GGCCAUC-------ACCCAACCAUUCCCC----UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACUCAACUCGACAUCGUCUGCCCCUCCUUGG -----(((....-------...............----..((((.(((((((((((.........)))))))))))))))........(((...))).)))......... ( -25.80) >DroSim_CAF1 30116 94 - 1 -----GGCCAUC-------ACCCAACAAUCCCCC----UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACUCGACUCGACAUCGUCUGCCCCUCUUUGG -----(((....-------...............----..((((.(((((((((((.........)))))))))))))))...(((........))).)))......... ( -27.30) >DroEre_CAF1 28509 93 - 1 CCAUCGCCCAUC-------ACCCAGCCAUCCCCC----UCUCGAAGUCAAGUGGCGCGCAACAAGUGCCACUUGACUCGACG-----CGACUUCGCAUAC-CCUCUUUGG ............-------..((((.........----..((((.((((((((((((.......)))))))))))))))).(-----((....)))....-.....)))) ( -32.70) >DroYak_CAF1 29387 79 - 1 ---------------------CCAGUCAUCCCCC----UCUCGAAGUCAAGUGGCGCGCAACAAGUGCUACUUGACUCGACU-----CGACUUCGUCUAC-CCUCUUUGG ---------------------((((.........----..((((.((((((((((((.......))))))))))))))))..-----.(((...)))...-.....)))) ( -26.30) >DroAna_CAF1 36273 92 - 1 -----CACCACCACAUCGCACACAACACUCGCACACAACCUCCAAGUCGAGUGCCAGGCAACAAGUGCCACUUGACACGACU-----UGGCUUCGUUG--------UUCA -----............((...........))..(((((..((((((((.((....((((.....)))).....)).)))))-----)))....))))--------)... ( -21.70) >consensus _____GGCCAUC_______ACCCAACCAUCCCCC____UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACU_____CGACAUCGUCUGC_CCUCUUUGG .....................((((...............((((.((((((((((((.......)))))))))))))))).......((....))...........)))) (-18.92 = -19.28 + 0.36)


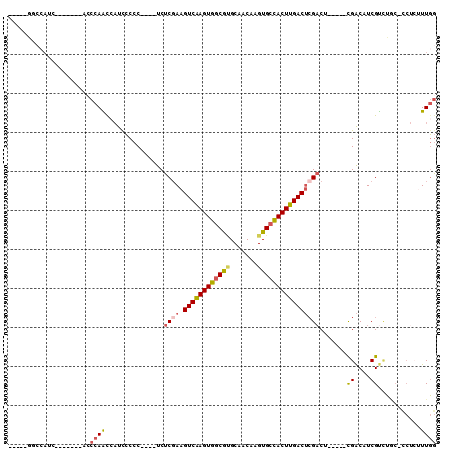
| Location | 3,343,237 – 3,343,332 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.86 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -17.05 |
| Energy contribution | -17.00 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3343237 95 - 22407834 AGUGGCAUCAGAUAAAUAGCACCCACCCAUCGCCU----------GGCCAUC-------ACCCAGCCAUCCCCC----UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGAC ((((((((..........((((((((.....(.((----------((.....-------..)))).).......----....(....)..)))).)))).....)))))))).... ( -23.66) >DroSec_CAF1 34067 95 - 1 AGUGGCAUCAGAUAAAUAGCACCCACCCAUCGCCC----------GGCCAUC-------ACCCAACCAUUCCCC----UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGAC .(((((....(((...............)))....----------.))))).-------...............----.......(((((((((((.........))))))))))) ( -22.56) >DroSim_CAF1 30150 95 - 1 AGUGGCAUCAGAUAAAUAGCACCCACCCAUCCCCC----------GGCCAUC-------ACCCAACAAUCCCCC----UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGAC .(((((....(((...............)))....----------.))))).-------...............----.......(((((((((((.........))))))))))) ( -21.46) >DroEre_CAF1 28537 105 - 1 AGUGGCAUCAGAUAAAUAGCACCCACCCAUCGCCCAUCGCCCAUCGCCCAUC-------ACCCAGCCAUCCCCC----UCUCGAAGUCAAGUGGCGCGCAACAAGUGCCACUUGAC .(((((............((...........)).....((.....)).....-------.....))))).....----.......((((((((((((.......)))))))))))) ( -27.60) >DroYak_CAF1 29415 77 - 1 AGUGGCAUCAGAUAAAUAGCACCCAC-----------------------------------CCAGUCAUCCCCC----UCUCGAAGUCAAGUGGCGCGCAACAAGUGCUACUUGAC .((((.................))))-----------------------------------.............----.......((((((((((((.......)))))))))))) ( -21.03) >DroAna_CAF1 36294 105 - 1 AGUGGCAUCAGAUAAACUGUC-UACCCCACCGCAC----------CACCACCACAUCGCACACAACACUCGCACACAACCUCCAAGUCGAGUGCCAGGCAACAAGUGCCACUUGAC ((((((((.((((.....)))-)............----------............((......((((((.((...........))))))))....)).....)))))))).... ( -22.70) >consensus AGUGGCAUCAGAUAAAUAGCACCCACCCAUCGCCC__________GGCCAUC_______ACCCAACCAUCCCCC____UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGAC .((((.................))))...........................................................((((((((((((.......)))))))))))) (-17.05 = -17.00 + -0.05)
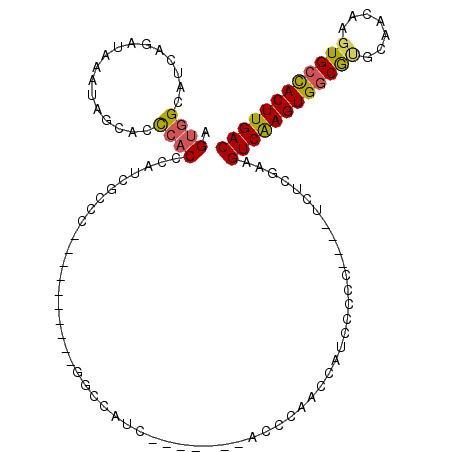
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:04 2006