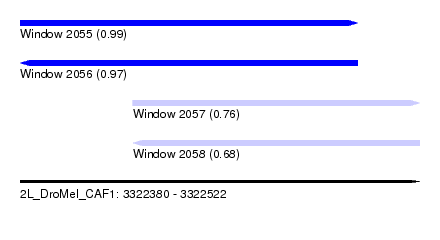
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,322,380 – 3,322,522 |
| Length | 142 |
| Max. P | 0.989175 |

| Location | 3,322,380 – 3,322,500 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -46.27 |
| Consensus MFE | -30.68 |
| Energy contribution | -31.88 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3322380 120 + 22407834 AUCUUUGGCUGCUGGGCAACUGGGCAGAUCGGUAUCUGCCCAUUAUUCUGCUGGCCAUUCAGCUGGCUCUUGUCAGCUUUGGCCUUUAUCCCCUGGCCGCAGUCGUUAGCAGCGAAGUUC ...(((.((((((((.(...(((((((((....))))))))).....((((.(((((...(((((((....)))))))..((........)).)))))))))..))))))))).)))... ( -51.60) >DroEre_CAF1 8250 120 + 1 ACCUUUGGCUGCUGGGCAACUGGGCGGAUCGCAAUUUGCCCAUUGUUCUGCUGGCCAUCCAGCUGGCUGUGGUCAGCUUUGGCCUUUAUCCCCUGGCCGCGGUUGUUAGCAGCGAGGUUU (((((..((((((((.(((((((((((((....))))))))........((((((((..(((....)))))))))))...((((..........))))..)))))))))))))))))).. ( -55.00) >DroWil_CAF1 118939 120 + 1 AAUCAUGGAUGUUGGGUUCGUGGUCGGAGCAAUAUCUUCCCAUUAUUUUACUGGCCGUGCAAUUGGCCUUUGUCAGCUUUGGGCUAUAUCCAUUGGCUGCUGUGGUAAGCAGCGAGCUUU ..((((((((((..(((((((((..(((......)))..)))).........(((((......)))))............)))))))))))))..((((((......))))))))..... ( -37.70) >DroYak_CAF1 8573 120 + 1 AAUUUUGGUUGCUGGGCAACUGGGCUGAUCGGUAUUUGCCCAUUGUUUUGCUGGCCAUCCAGCUGGCUGUGGUCAGCUUUGGCCUUUAUCCCCUGGCCGCAGUUGUUAGCAGCGAGGUUC ...(((.((((((((.(((((((((((((.(((....))).)))........(((((...(((((((....))))))).)))))..........)))).)))))))))))))).)))... ( -49.00) >DroMoj_CAF1 9397 120 + 1 AGAGCUGGUCUCUGGGCCCCUGGAGUGAGCGCACUGUUCCCAUCAUCCUGCUCGCCAUUCAGCUAUUCUUCGUCAGCUUUGGGCUCUAUCCCCUGGCCGCUGUCGUCAGCAGCGAAGUGC ..((((((..((..(....)..))(((((((...((.......))...)))))))...))))))...((((((..(((..(((......)))..))).((((....)))).))))))... ( -39.10) >DroAna_CAF1 8475 120 + 1 AAAGUUGGCUGCUGGGUGAAUGGGCGGAUCGGUAUUUGCCCAUUAUCCUAUUGGCCAUCCAACUCGCCUUUGUUAGUUUCGGUCUAUAUCCACUGGCAGCUGUGGUGAGCAGCGAGGUUU (((.((.((((((((((((((((((((((....)))))))))))..((....)).))))))..(((((.((((((((...((.......))))))))))....)))))))))).)).))) ( -45.20) >consensus AAAUUUGGCUGCUGGGCAACUGGGCGGAUCGGUAUUUGCCCAUUAUUCUGCUGGCCAUCCAGCUGGCCUUUGUCAGCUUUGGCCUUUAUCCCCUGGCCGCUGUCGUUAGCAGCGAGGUUC .......((((((((((((.(((((((((....)))))))))))))))...((((((...(((((((....)))))))..((.......))..))))))........))))))....... (-30.68 = -31.88 + 1.20)


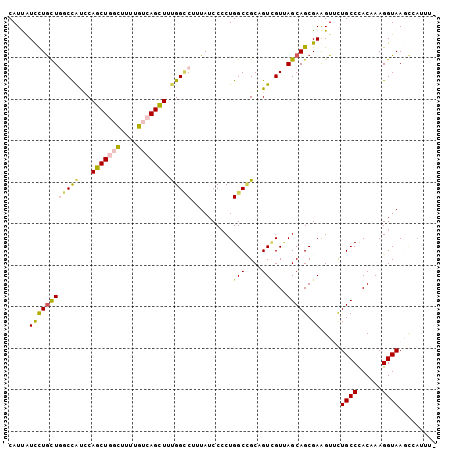
| Location | 3,322,380 – 3,322,500 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -21.29 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3322380 120 - 22407834 GAACUUCGCUGCUAACGACUGCGGCCAGGGGAUAAAGGCCAAAGCUGACAAGAGCCAGCUGAAUGGCCAGCAGAAUAAUGGGCAGAUACCGAUCUGCCCAGUUGCCCAGCAGCCAAAGAU ...(((.((((((.....(((....)))((......(((((.(((((........)))))...))))).((((.....(((((((((....))))))))).))))))))))))..))).. ( -49.00) >DroEre_CAF1 8250 120 - 1 AAACCUCGCUGCUAACAACCGCGGCCAGGGGAUAAAGGCCAAAGCUGACCACAGCCAGCUGGAUGGCCAGCAGAACAAUGGGCAAAUUGCGAUCCGCCCAGUUGCCCAGCAGCCAAAGGU ..((((.((((((.....((.(.....).)).....(((((.(((((........)))))...)))))...........((((((...(((...)))....))))))))))))...)))) ( -44.90) >DroWil_CAF1 118939 120 - 1 AAAGCUCGCUGCUUACCACAGCAGCCAAUGGAUAUAGCCCAAAGCUGACAAAGGCCAAUUGCACGGCCAGUAAAAUAAUGGGAAGAUAUUGCUCCGACCACGAACCCAACAUCCAUGAUU .......((((((......))))))..((((((.((((.....)))).....((((........))))..........((((..((......))((....))..))))..)))))).... ( -29.10) >DroYak_CAF1 8573 120 - 1 GAACCUCGCUGCUAACAACUGCGGCCAGGGGAUAAAGGCCAAAGCUGACCACAGCCAGCUGGAUGGCCAGCAAAACAAUGGGCAAAUACCGAUCAGCCCAGUUGCCCAGCAACCAAAAUU ...(((.(((((........))))).)))((.....(((((.(((((........)))))...))))).((((.....(((((............))))).)))).......))...... ( -42.50) >DroMoj_CAF1 9397 120 - 1 GCACUUCGCUGCUGACGACAGCGGCCAGGGGAUAGAGCCCAAAGCUGACGAAGAAUAGCUGAAUGGCGAGCAGGAUGAUGGGAACAGUGCGCUCACUCCAGGGGCCCAGAGACCAGCUCU ...(((((((((((....)))))))((((((......)))....)))..))))....(((....)))((((.((.(..((((..(((((....))))....)..))))..).)).)))). ( -40.30) >DroAna_CAF1 8475 120 - 1 AAACCUCGCUGCUCACCACAGCUGCCAGUGGAUAUAGACCGAAACUAACAAAGGCGAGUUGGAUGGCCAAUAGGAUAAUGGGCAAAUACCGAUCCGCCCAUUCACCCAGCAGCCAACUUU .......((((((..(((((((((((..((....(((.......))).))..))).)))))..)))......((..(((((((............)))))))...))))))))....... ( -32.90) >consensus AAACCUCGCUGCUAACAACAGCGGCCAGGGGAUAAAGGCCAAAGCUGACAAAAGCCAGCUGGAUGGCCAGCAGAAUAAUGGGCAAAUACCGAUCCGCCCAGUUGCCCAGCAGCCAAAAUU ...(((.(((((........))))).)))((.....(.(((.(((((........)))))...))).)..........(((((............))))).....))............. (-21.29 = -21.77 + 0.48)
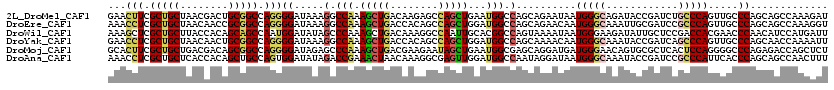
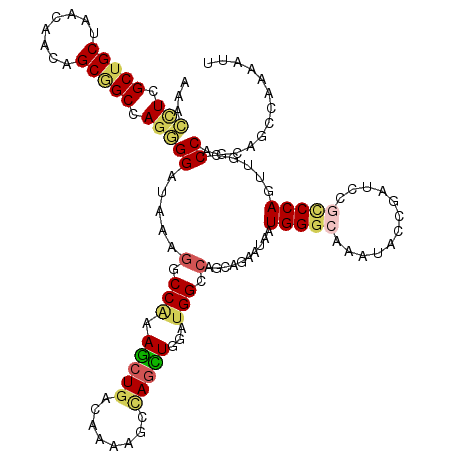
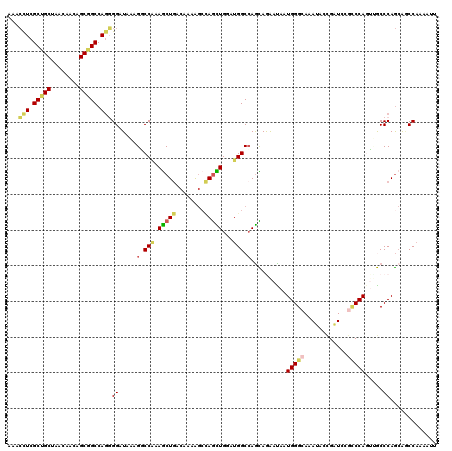
| Location | 3,322,420 – 3,322,522 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.82 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3322420 102 + 22407834 CAUUAUUCUGCUGGCCAUUCAGCUGGCUCUUGUCAGCUUUGGCCUUUAUCCCCUGGCCGCAGUCGUUAGCAGCGAAGUUCUGCCCACGAAGGUAAUCCAUUA- .......((((.(((((...(((((((....)))))))..((........)).)))))))))((((..((((.......))))..)))).((....))....- ( -32.70) >DroVir_CAF1 32054 102 + 1 CAUCGUUCUGCUCGCCGCUCAGCUGUUUUUUGUCAGCUUUGGUCUCUAUCCCCUGGCUGCUGUCGUCAGCAGCGAAGUGCUGCCCACAAAGGUAAGGAAUAU- ....(((((((.(((((...(((((........))))).))))............(((((((....)))))))...).))((((......)))).)))))..- ( -29.60) >DroSim_CAF1 8737 102 + 1 CAUUGUCCUGCUGGCUAUCCAGCUGGCUCUUGUCAGCUUUGGCCUUUAUCCCCUGGCCGCAGUCGUUAGCAGCGAAGUUCUGCCCACAAAGGUAAUCCAUUA- .....((((((((((.....(((((((....)))))))..((((..........))))......)))))))).)).....((((......))))........- ( -33.00) >DroYak_CAF1 8613 102 + 1 CAUUGUUUUGCUGGCCAUCCAGCUGGCUGUGGUCAGCUUUGGCCUUUAUCCCCUGGCCGCAGUUGUUAGCAGCGAGGUUCUGCCCACUAAGGUAAGCCAGUU- .........((((((.......((.(((((...(((((.(((((..........))))).)))))...))))).))....((((......)))).)))))).- ( -38.70) >DroMoj_CAF1 9437 101 + 1 CAUCAUCCUGCUCGCCAUUCAGCUAUUCUUCGUCAGCUUUGGGCUCUAUCCCCUGGCCGCUGUCGUCAGCAGCGAAGUGCUGCCCACAAAGGUAAGCAAUA-- ........((((..((.((((((....((((((..(((..(((......)))..))).((((....)))).)))))).)))).....)).))..))))...-- ( -31.90) >DroAna_CAF1 8515 102 + 1 CAUUAUCCUAUUGGCCAUCCAACUCGCCUUUGUUAGUUUCGGUCUAUAUCCACUGGCAGCUGUGGUGAGCAGCGAGGUUUUGCCUACAAAGGUAAGUUU-UUG .............(((.(((..((((((.((((((((...((.......))))))))))....))))))..).)))))(((((((....)))))))...-... ( -25.10) >consensus CAUUAUCCUGCUGGCCAUCCAGCUGGCUUUUGUCAGCUUUGGCCUUUAUCCCCUGGCCGCAGUCGUUAGCAGCGAAGUUCUGCCCACAAAGGUAAGCCAUUU_ .....((((((((((((...(((((((....))))))).)))))..........(((....)))...))))).)).....((((......))))......... (-20.70 = -20.82 + 0.12)

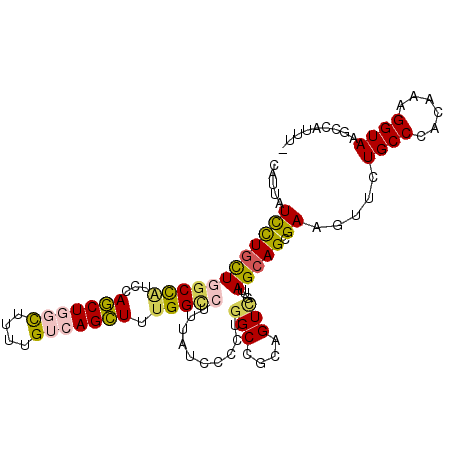
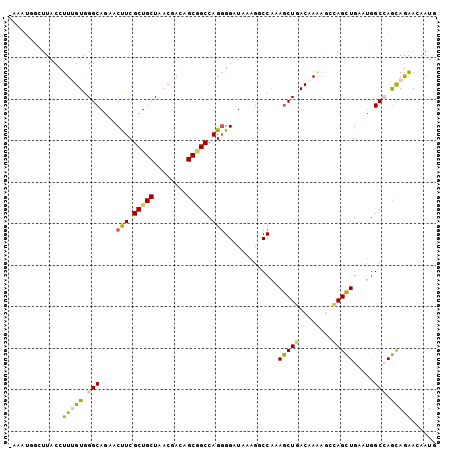
| Location | 3,322,420 – 3,322,522 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -20.83 |
| Energy contribution | -20.62 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3322420 102 - 22407834 -UAAUGGAUUACCUUCGUGGGCAGAACUUCGCUGCUAACGACUGCGGCCAGGGGAUAAAGGCCAAAGCUGACAAGAGCCAGCUGAAUGGCCAGCAGAAUAAUG -.............((((.(((((.......))))).))))(((((((((.((........))..(((((........)))))...))))).))))....... ( -33.40) >DroVir_CAF1 32054 102 - 1 -AUAUUCCUUACCUUUGUGGGCAGCACUUCGCUGCUGACGACAGCAGCCAGGGGAUAGAGACCAAAGCUGACAAAAAACAGCUGAGCGGCGAGCAGAACGAUG -.............((((.((((((.....)))))).))))..((.(((..((........))..(((((........)))))....)))..))......... ( -30.40) >DroSim_CAF1 8737 102 - 1 -UAAUGGAUUACCUUUGUGGGCAGAACUUCGCUGCUAACGACUGCGGCCAGGGGAUAAAGGCCAAAGCUGACAAGAGCCAGCUGGAUAGCCAGCAGGACAAUG -..........(((((((.(((....(((.(((((........))))).)))((.......))...))).))))......(((((....))))))))...... ( -30.20) >DroYak_CAF1 8613 102 - 1 -AACUGGCUUACCUUAGUGGGCAGAACCUCGCUGCUAACAACUGCGGCCAGGGGAUAAAGGCCAAAGCUGACCACAGCCAGCUGGAUGGCCAGCAAAACAAUG -...(((((....(((((.(((....(((.(((((........))))).)))........)))...)))))....)))))(((((....)))))......... ( -37.00) >DroMoj_CAF1 9437 101 - 1 --UAUUGCUUACCUUUGUGGGCAGCACUUCGCUGCUGACGACAGCGGCCAGGGGAUAGAGCCCAAAGCUGACGAAGAAUAGCUGAAUGGCGAGCAGGAUGAUG --..((((((..((((((.(((....((..(((((((....))))))).))(((......)))...))).))))))....(((....)))))))))....... ( -36.40) >DroAna_CAF1 8515 102 - 1 CAA-AAACUUACCUUUGUAGGCAAAACCUCGCUGCUCACCACAGCUGCCAGUGGAUAUAGACCGAAACUAACAAAGGCGAGUUGGAUGGCCAAUAGGAUAAUG ((.-.((((..(((((((.((((.......((((.......))))))))..(((.......)))......)))))))..))))...)).((....))...... ( -23.61) >consensus _AAAUGGCUUACCUUUGUGGGCAGAACUUCGCUGCUAACGACAGCGGCCAGGGGAUAAAGGCCAAAGCUGACAAAAGCCAGCUGAAUGGCCAGCAGAACAAUG .............(((((.(((....(((.(((((........))))).))).............(((((........))))).....))).)))))...... (-20.83 = -20.62 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:57 2006