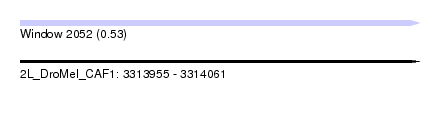
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,313,955 – 3,314,061 |
| Length | 106 |
| Max. P | 0.533604 |

| Location | 3,313,955 – 3,314,061 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -15.70 |
| Energy contribution | -16.23 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3313955 106 + 22407834 ---UAUGCUU--UUAGGCACUUCGUACAAUGUUCUUGUAAGCGCGGUGAACAACAGUCGUUGUCCGGAUGCGCCGUGCAGCGAGUACGGAAUCAAAGAAAAGAAUGCAUCU ---..((((.--...)))).(((((((..(((((((((....)))).)))))....((((((..(((.....)))..)))))))))))))..................... ( -30.90) >DroVir_CAF1 21195 97 + 1 ------CCCA--ACAGGCACCACAUACAAUGUGUUGGUGAGCGCUGUGAAUAGCACUCGCUGCCCGGAUGCGCCCUGCAGCGAGUACACCAUCAAACAGAAGC------CC ------....--...(((..(((((...))))).(((((...((((....))))(((((((((..((.....))..))))))))).)))))..........))------). ( -35.50) >DroPse_CAF1 24826 98 + 1 -----CUUUU--GCAGGCACUUCCUACAAUGUCCUUGUCAGUGCCGUGAACAACACACGCUGUCCGGAUGCCCCCUGCAGCGAGUACGGCAUCAAGGAGAAGC------CA -----.....--...(((.....(......)((((((...(((((((.......((.((((((..((.....))..)))))).)))))))))))))))...))------). ( -31.81) >DroWil_CAF1 111252 100 + 1 -----UUGUUGCACAGGCACUUCCUACAACGUGCUUGUCAGUGCUGUGAACAACACACGCUGUCCGGAUGCCCCGUGCAGCGAGUACGGCAUCAAAGAGAAGC------CA -----....((.((((((((..........))))))))))((((((((.....))).(((((..(((.....)))..))))))))))(((.((.....)).))------). ( -39.00) >DroAna_CAF1 162 103 + 1 UAUUUUACAU--UCAGGCACUUCAUACAAUGUCCUUGUGAGUGCUCUGAAUAACAGUCGGUGUCCGGAUGCGCCGUGCAGCGAAUAUGGCAUCAAACAGAAGC------CC ........((--((((((((((...((((.....)))))))))).)))))).....(((.((..(((.....)))..)).)))....(((.((.....)).))------). ( -31.80) >DroPer_CAF1 27004 98 + 1 -----CUUUU--GCAGGCACUUCCUACAAUGUCCUUGUCAGUGCCGUGAACAACACACGCUGUCCGGAUGCCCCCUGCAGCGAGUACGGCAUCAAGGAGAAGC------CA -----.....--...(((.....(......)((((((...(((((((.......((.((((((..((.....))..)))))).)))))))))))))))...))------). ( -31.81) >consensus _____UUCUU__ACAGGCACUUCCUACAAUGUCCUUGUCAGUGCUGUGAACAACACACGCUGUCCGGAUGCCCCCUGCAGCGAGUACGGCAUCAAAGAGAAGC______CA ...............((((((....((((.....)))).))))))(((.....))).((((((..((.....))..))))))............................. (-15.70 = -16.23 + 0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:51 2006