
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,311,324 – 3,311,524 |
| Length | 200 |
| Max. P | 0.899367 |

| Location | 3,311,324 – 3,311,444 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -47.80 |
| Consensus MFE | -41.43 |
| Energy contribution | -42.07 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
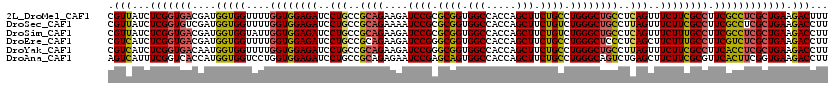
>2L_DroMel_CAF1 3311324 120 + 22407834 CAGAAGGUGGACCUAUCGGCCUUCUCCGCCUGGCUGCCCGUCACCCUAAUGGCCUUCAUCAUCUUCAUUGCCAACGUGGGCGUUAUCUCGGUGACGAUGGUGGUUUUGGUGGAGAUCCUG (((.((((.((....)).))))((((((((.(((..(((((((((.....(((................)))((((....)))).....)))))))..))..)))..))))))))..))) ( -46.19) >DroSec_CAF1 20245 120 + 1 CAGAAGGCGGACCUGUCGGCCUUCUCCGCCUGGCUGCCCGUCACCCUGAUGGCCUUCAUCAUCUUCAUUGCCAACGUGGGCGUUAUCUCGGUGUCGAUGGUGGUUUUGGUGGAGAUCCUG (((.((((.((....)).))))((((((((.(((..((((.((((.(((((.....)))))........(((......)))........)))).))..))..)))..))))))))..))) ( -48.10) >DroSim_CAF1 20219 120 + 1 CAGAAGGUGGACCUGUCGGCCUUCUCCGCCUGGCUGCCCGUCACCCUGAUGGCCUUCAUCAUCUUCAUUGCCAACGUGGGCGUUAUCUCGGUGACGAUGGUGGUAUUGGUGGAGAUCCUG (((.((((.((....)).))))((((((((..((..(((((((((.(((((.....)))))........(((......)))........)))))))..))..))...))))))))..))) ( -51.00) >DroEre_CAF1 28463 120 + 1 CAGAAGGUGGACCUGUCGGCCUUCUCCGCCUGGCUGCCCGUCACCCUGAUGGCCUUCAUCAUCUUCAUUGCCAACGUGGGCGUCAUCUCGGUGACGAUGGUGGUUUUGGUGGAGAUCCUG (((.((((.((....)).))))((((((((.(((..(((((((((..((((((.(((((................))))).))))))..)))))))..))..)))..))))))))..))) ( -52.79) >DroYak_CAF1 20504 120 + 1 GAGAAGGCGGACCUAUCGGCCUUUUCCGCCUGGCUACCCGUCACCCUGAUGGCCUUUAUCAUCUUCAUUGCCAACGUGGGCGUCAUCUCGGUGACAAUGGUGGUUUUGGUGGAGAUCCUG ..((((((.((....)).))))))((((((.(((((((.((((((..((((((................(((......)))))))))..))))))...)))))))..))))))....... ( -47.99) >DroAna_CAF1 30184 120 + 1 CUGGAAGCAGACAUGUCUGCCUACUCCGCGUGGCUGCCCGUGUGCCUCAUGUCCUUCAUUAUAUUCAUUGCCAACGUGGGAGUCAUUUCGGUCACCAUGGUGGUCCUGGUGGAGAUCCUG ..(((.(((((....)))))...((((((((((((.(((((..((...(((..............))).))..))).)).))))))...(..(((....)))..)...)))))).))).. ( -40.74) >consensus CAGAAGGCGGACCUGUCGGCCUUCUCCGCCUGGCUGCCCGUCACCCUGAUGGCCUUCAUCAUCUUCAUUGCCAACGUGGGCGUCAUCUCGGUGACGAUGGUGGUUUUGGUGGAGAUCCUG ..((((((.((....)).))))))((((((..(((((((((((((..((((((.(((((................))))).))))))..)))))))..))))))...))))))....... (-41.43 = -42.07 + 0.64)


| Location | 3,311,404 – 3,311,524 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.89 |
| Mean single sequence MFE | -47.82 |
| Consensus MFE | -40.74 |
| Energy contribution | -40.27 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3311404 120 + 22407834 CGUUAUCUCGGUGACGAUGGUGGUUUUGGUGGAGAUCCUGCCGCAGAAGAUCCGCGCGGUGGCCACCAGCUUCUGCCUGGGCUGCCUCAGUUUCUUCGCCUUCGCCUCGCUGAAGACUUU .....(((((((((....(((((....((((((((..(((..((((....((((.((((.(((.....))).)))).))))))))..)))..)))))))).))))))))))).))).... ( -48.00) >DroSec_CAF1 20325 120 + 1 CGUUAUCUCGGUGUCGAUGGUGGUUUUGGUGGAGAUCCUGCCGCAGAAAAUCCGCGCGGUGGCCACCAGCUUCUGUCUGGGCUGCCUUAGUUUCUUCGCCUUCGCCUCGCUGAAGACCUU .....((((((((.....(((((....((((((((..(((((((.(......)..)))))(((..(((((....).))))...)))..))..)))))))).))))).))))).))).... ( -44.60) >DroSim_CAF1 20299 120 + 1 CGUUAUCUCGGUGACGAUGGUGGUAUUGGUGGAGAUCCUGCCGCAGAAGAUCCGCGCGGUGGCCACCAGCUUCUGUCUGGGCUGCCUCAGUUUCUUUGCCUUCGCCUCGCUGAAGACCUU .....(((((((((....(((((....((..((((..(((..((((....((((.((((.(((.....))).)))).))))))))..)))..))))..)).))))))))))).))).... ( -45.00) >DroEre_CAF1 28543 120 + 1 CGUCAUCUCGGUGACGAUGGUGGUUUUGGUGGAGAUCCUGCCGCAGAAGAUCCGGGCGGUGGCCACCAGCUUCUGCCUGGGCUCCCUCAGCUUCUUUGCCUUCGUCUCGCUGAAGACCUU .(((...(((((((.(((((.(((...((..(.....)..))...(((((.((((((((.(((.....))).))))))))(((.....))).)))))))).)))))))))))).)))... ( -47.00) >DroYak_CAF1 20584 120 + 1 CGUCAUCUCGGUGACAAUGGUGGUUUUGGUGGAGAUCCUGCCGCAGAAGAUCCGGGCGGUGGCCACCAGCUUCUGCCUGGGCUGCCUUAGUUUCUUCGCCUUCACCUCGCUGAAGACCUU .(((...(((((((....(((((....((((((((..(((..((((....(((((((((.(((.....))).)))))))))))))..)))..)))))))).)))))))))))).)))... ( -54.60) >DroAna_CAF1 30264 120 + 1 AGUCAUUUCGGUCACCAUGGUGGUCCUGGUGGAGAUCCUGCCGCAGAGAAUCCGAGCAGUGGCCACCAGCUUCUGCCUGGGCAGUCUGAGCUUCUUCGCGUUCACUUCGGUGAAGACCUU .........((((....(((((((((((.((((..(((((...))).)).))))..))).))))))))((((((((....))))...)))).........(((((....))))))))).. ( -47.70) >consensus CGUCAUCUCGGUGACGAUGGUGGUUUUGGUGGAGAUCCUGCCGCAGAAGAUCCGCGCGGUGGCCACCAGCUUCUGCCUGGGCUGCCUCAGUUUCUUCGCCUUCGCCUCGCUGAAGACCUU .(((...(((((((....(((((....((((((((..(((..((((....((((.((((.(((.....))).)))).))))))))..)))..)))))))).)))))))))))).)))... (-40.74 = -40.27 + -0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:49 2006