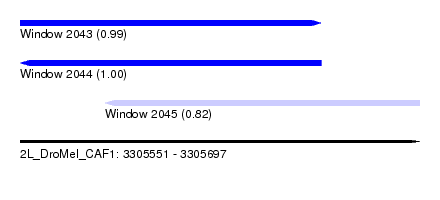
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,305,551 – 3,305,697 |
| Length | 146 |
| Max. P | 0.998440 |

| Location | 3,305,551 – 3,305,661 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -18.24 |
| Consensus MFE | -9.50 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3305551 110 + 22407834 AACAA-CACAAGUAU--------AGUAUCAAUAUAUGUAUAUAUACAUAUAUACGCUAUAUGCCCU-GCACAUGAUUAAGUAGCCCCAUUUAAUCUACUUAAACACGCGCCCCAAUUGAU .....-..(((((((--------(((....(((((((((....)))))))))..))))))).....-((.(.((.((((((((...........)))))))).)).).)).....))).. ( -21.80) >DroSim_CAF1 14604 105 + 1 AACAA-CACAAGUAC--------AGUAUCAAUAUAUAUAU------AUAUAUACGCUGCAGGGCAUAGCACAUGAUUAAGUAGCCCCAUUUAAUCUACUUAAACACGCGCCCCAAUUGAU .....-..(((...(--------(((....((((((....------))))))..))))..((((...((...((.((((((((...........)))))))).)).))))))...))).. ( -18.10) >DroEre_CAF1 22733 100 + 1 AACGA-CACAAGUAC--------AGUAUCAAUACA-------G--UAUAUAUACGCUAUAUAC-CA-GCACAUGAUUAAGUAGCCCCAUUUAAUCUACUUAAACACGCGCCCCAAUUGAU .....-.........--------...((((((...-------(--((((((.....)))))))-..-((.(.((.((((((((...........)))))))).)).).))....)))))) ( -16.60) >DroYak_CAF1 14738 108 + 1 AACCA-CACAAGUAC--------AGUAUCAAUAUAUAUACAUA--UACAUAUACGCUAUAUACCCA-GCACAUGAUUAAGUAGCCCCAUUUAAUCUACUUAAACACGCGCACCAAUUGAU .....-.........--------...((((((.(((((((.((--(....))).).))))))....-((.(.((.((((((((...........)))))))).)).).))....)))))) ( -14.50) >DroAna_CAF1 24654 107 + 1 UACAAAGGCCAAUACUUUCGAGGAGUAUCGAGCCG-------G--AAUAAA---AGUAUAUACCCU-GCACAUGAUUAAGUAGCCUCAUUUAAUCUACUUAAACACGCGCUAAAAUAAAU ......((((.((((((.....)))))).).)))(-------(--.(((..---....))).))..-((.(.((.((((((((...........)))))))).)).).)).......... ( -20.20) >consensus AACAA_CACAAGUAC________AGUAUCAAUAUAU_UA___A__UAUAUAUACGCUAUAUACCCU_GCACAUGAUUAAGUAGCCCCAUUUAAUCUACUUAAACACGCGCCCCAAUUGAU ...................................................................((.(.((.((((((((...........)))))))).)).).)).......... ( -9.50 = -9.50 + 0.00)

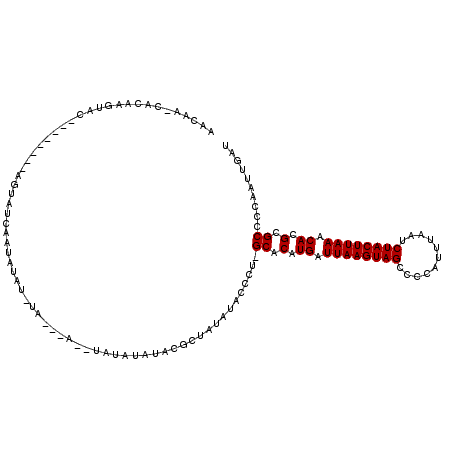
| Location | 3,305,551 – 3,305,661 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.60 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3305551 110 - 22407834 AUCAAUUGGGGCGCGUGUUUAAGUAGAUUAAAUGGGGCUACUUAAUCAUGUGC-AGGGCAUAUAGCGUAUAUAUGUAUAUAUACAUAUAUUGAUACU--------AUACUUGUG-UUGUU ..........(((((((.((((((((...........)))))))).)))))))-((.((((((((..((((((((((....)))))))))..)..))--------)))..))).-))... ( -29.40) >DroSim_CAF1 14604 105 - 1 AUCAAUUGGGGCGCGUGUUUAAGUAGAUUAAAUGGGGCUACUUAAUCAUGUGCUAUGCCCUGCAGCGUAUAUAU------AUAUAUAUAUUGAUACU--------GUACUUGUG-UUGUU ..((((.((((((((((.((((((((...........)))))))).)))))))....)))(((((.(((((((.------...))))))).....))--------))).....)-))).. ( -26.50) >DroEre_CAF1 22733 100 - 1 AUCAAUUGGGGCGCGUGUUUAAGUAGAUUAAAUGGGGCUACUUAAUCAUGUGC-UG-GUAUAUAGCGUAUAUAUA--C-------UGUAUUGAUACU--------GUACUUGUG-UCGUU .........((((((((.((((((((...........)))))))).)))))))-)(-(((((((.....))))))--)-------).....(((((.--------......)))-))... ( -30.50) >DroYak_CAF1 14738 108 - 1 AUCAAUUGGUGCGCGUGUUUAAGUAGAUUAAAUGGGGCUACUUAAUCAUGUGC-UGGGUAUAUAGCGUAUAUGUA--UAUGUAUAUAUAUUGAUACU--------GUACUUGUG-UGGUU ((((((....(((((((.((((((((...........)))))))).)))))))-...(((((((.(((((....)--)))))))))))))))))((.--------.((....))-..)). ( -29.50) >DroAna_CAF1 24654 107 - 1 AUUUAUUUUAGCGCGUGUUUAAGUAGAUUAAAUGAGGCUACUUAAUCAUGUGC-AGGGUAUAUACU---UUUAUU--C-------CGGCUCGAUACUCCUCGAAAGUAUUGGCCUUUGUA ..........(((((((.((((((((.((....))..)))))))).)))))))-.(((.(((....---..))))--)-------)(((.(((((((.......))))))))))...... ( -32.60) >consensus AUCAAUUGGGGCGCGUGUUUAAGUAGAUUAAAUGGGGCUACUUAAUCAUGUGC_AGGGUAUAUAGCGUAUAUAUA__C___UA_AUAUAUUGAUACU________GUACUUGUG_UUGUU ((((((....(((((((.((((((((...........)))))))).))))))).....((((((.....)))))).............)))))).......................... (-17.74 = -18.50 + 0.76)

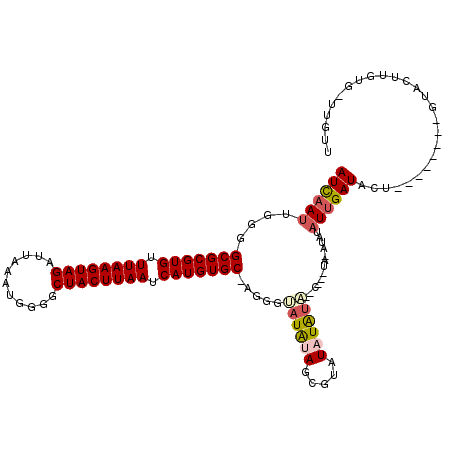
| Location | 3,305,582 – 3,305,697 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.65 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -20.62 |
| Energy contribution | -22.22 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.816034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
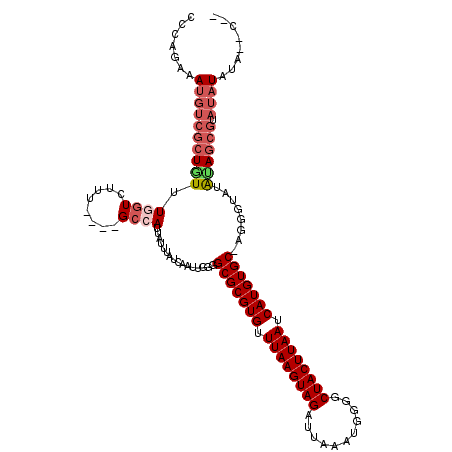
>2L_DroMel_CAF1 3305582 115 - 22407834 CCCAGAAAUGUCGCUAUUUGGUCUUU----GCCAUUAUUUAUCAAUUGGGGCGCGUGUUUAAGUAGAUUAAAUGGGGCUACUUAAUCAUGUGC-AGGGCAUAUAGCGUAUAUAUGUAUAU .......((((((((((...(((((.----.(((............))).(((((((.((((((((...........)))))))).)))))))-)))))..)))))).))))........ ( -33.40) >DroSim_CAF1 14635 110 - 1 CCCAGAAAUGUCGCUGUUUAGUCUUU----GCCAUUAUUUAUCAAUUGGGGCGCGUGUUUAAGUAGAUUAAAUGGGGCUACUUAAUCAUGUGCUAUGCCCUGCAGCGUAUAUAU------ .......((((((((((...((....----.(((............)))((((((((.((((((((...........)))))))).))))))))..))...)))))).))))..------ ( -31.50) >DroEre_CAF1 22759 114 - 1 CCCAGAAAUGUCGCUGUUUGGUCUUUGACUGCCAUUAUUUAUCAAUUGGGGCGCGUGUUUAAGUAGAUUAAAUGGGGCUACUUAAUCAUGUGC-UG-GUAUAUAGCGUAUAUAUA--C-- ((((((((((..((.(((........))).))...))))).....)))))(((((((.((((((((...........)))))))).)))))))-..-(((((((.....))))))--)-- ( -30.60) >DroYak_CAF1 14769 117 - 1 CCCAGAAAUGUCGCUGUUUGGUCUUUGACUGCCAUUAUUUAUCAAUUGGUGCGCGUGUUUAAGUAGAUUAAAUGGGGCUACUUAAUCAUGUGC-UGGGUAUAUAGCGUAUAUGUA--UAU .......((((((((((.((.((.......((((............))))(((((((.((((((((...........)))))))).)))))))-.)).)).)))))).))))...--... ( -31.50) >DroAna_CAF1 24689 107 - 1 UCCAAAAAUUUC-CUUUUUCUUUUCU----GCUAUUAUUUAUUUAUUUUAGCGCGUGUUUAAGUAGAUUAAAUGAGGCUACUUAAUCAUGUGC-AGGGUAUAUACU---UUUAUU--C-- ............-.............----.........(((.((((((.(((((((.((((((((.((....))..)))))))).)))))))-)))))).)))..---......--.-- ( -20.70) >consensus CCCAGAAAUGUCGCUGUUUGGUCUUU____GCCAUUAUUUAUCAAUUGGGGCGCGUGUUUAAGUAGAUUAAAUGGGGCUACUUAAUCAUGUGC_AGGGUAUAUAGCGUAUAUAUA__C__ .......((((((((((.((((........))))................(((((((.((((((((...........)))))))).)))))))........)))))).))))........ (-20.62 = -22.22 + 1.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:44 2006