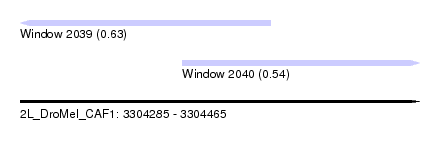
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,304,285 – 3,304,465 |
| Length | 180 |
| Max. P | 0.629972 |

| Location | 3,304,285 – 3,304,398 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -18.12 |
| Consensus MFE | -0.58 |
| Energy contribution | -0.86 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.03 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3304285 113 - 22407834 AUAUGAUUUGCAUUGCAAUUUAUUCAAUUUCCGUUUCCGCUUCUUUCACGCG-------GAACACCGAACGCACUUUGGACAAACUUGCACUAAAAAUAUUAUGCAUCAAUUUUAAUUUC ...(((..(((((((((((((.(((((....(((((((((.........)))-------))......))))....))))).))).)))))...........))))))))........... ( -20.00) >DroSec_CAF1 13147 113 - 1 AUAUGAUUUGCAUUGUAAUUUAUUCCAUUUUCGUUUCCGCUUCUUUCGCGCG-------GAACACCGAACGCACUUUGCACAAACUUGCACUAAAAAUAUUAUGCAUCAAUUUAAAUUUC ...(((..(((((.(((.(((........((((.((((((.........)))-------)))...)))).......((((......))))...))).))).))))))))........... ( -19.50) >DroSim_CAF1 13338 113 - 1 AUAUGAUUUGCAUUGUAAUUUAUUCCAUUUCCGUUUCCGCUUCUUUCGCGCG-------GAACACCGAACGCACUUUGCACAAACUUGCACCAAAAAUAUUAUGAAUCAAUUUAAAUUUC ....((((((.((((..((((((.....(((.((((((((.........)))-------))).)).))).......((((......))))...........)))))))))).)))))).. ( -17.10) >DroEre_CAF1 21561 120 - 1 AUCUGAUAUGCACUUUAAUUUAUUGCAUUUCCGUUUCCGCUUCGCACACGCAGAAUACCGAAUACCGAACUCGCUUUGUAUGCACUCGCACUUAUAAUAUUAUGCAUCAAUAUUAAUUCC ....((.(((((...........))))).)).......((...(((.(((.((.....((.....))......)).))).)))....)).....(((((((.......)))))))..... ( -12.40) >DroYak_CAF1 13439 102 - 1 UUAUGAUAUGCAGAUUAAUUUGUUGCAUUUCCGUUUCCGC-----------A-------GGAUAACAAACUUACUCUGCAUGCACAUGCACUAAUAAUAUUAUGAUUUUAUAUUAACUCC ((((((((((((((.((((((((((....(((........-----------.-------)))))))))).))).))))).(((....)))......)))))))))............... ( -21.60) >consensus AUAUGAUUUGCAUUGUAAUUUAUUCCAUUUCCGUUUCCGCUUCUUUCACGCG_______GAACACCGAACGCACUUUGCACAAACUUGCACUAAAAAUAUUAUGCAUCAAUUUUAAUUUC ............................................................................((((......)))).............................. ( -0.58 = -0.86 + 0.28)

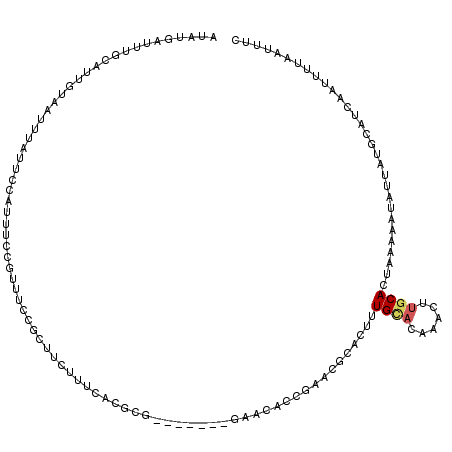
| Location | 3,304,358 – 3,304,465 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.87 |
| Mean single sequence MFE | -18.86 |
| Consensus MFE | -5.44 |
| Energy contribution | -6.44 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3304358 107 + 22407834 GCGGAAACGGAAAUUGAAUAAAUUGCAAUGCAAAUCAUAUGCAUAAAGGGAG----UGAGCCGU---------UAAUUUUGGUGAAAAAGUGAAGGAAAUCCUAAAAAAUGCAAAUCAAG .((....)).............((((((((((.......)))))...((((.----....((..---------..(((((......)))))...))...))))......)))))...... ( -20.80) >DroSec_CAF1 13220 107 + 1 GCGGAAACGAAAAUGGAAUAAAUUACAAUGCAAAUCAUAUGCAUAAAGGGAG----UGACCCGU---------UAAUUAUGAUGAAAGAGUGAAGGAAAUCCUAAAUAAUGCAAAUCAAG .((....))...................((((.((((((....(((.(((..----...))).)---------))..))))))..........(((....)))......))))....... ( -18.10) >DroSim_CAF1 13411 111 + 1 GCGGAAACGGAAAUGGAAUAAAUUACAAUGCAAAUCAUAUGCAUAAAGGGAGUGAGUGACCCGU---------UAAUUAUGGUGAAAGAGUGAAGGAAAUCCUAAAUAAUGAAAAUCAAG .((....))....(((......((((.(((((.......)))))...(((.........)))..---------................))))(((....)))............))).. ( -15.50) >DroEre_CAF1 21641 100 + 1 GCGGAAACGGAAAUGCAAUAAAUUAAAGUGCAUAUCAGAUGGCCCAACU--------------------AGUGUAAUUAUGGUGAACGAAUCAAGGAAAUCAUUAAUAAUGGAAAUCAAA .((....))((.(((((...........))))).)).(((..((....(--------------------((((......((((......)))).......))))).....))..)))... ( -17.02) >DroYak_CAF1 13501 113 + 1 GCGGAAACGGAAAUGCAACAAAUUAAUCUGCAUAUCAUAAGCAUCAAUGAAA----GUGCCCGUUGAAUGGUGAA---AUGGUGAAAAAAUCACGGAAAUCAUUAAUAAUGGAAAUCAAA .((....))((.(((((...........))))).))....((((........----))))((((((((((((...---.(.((((.....)))).)..))))))..))))))........ ( -22.90) >consensus GCGGAAACGGAAAUGGAAUAAAUUACAAUGCAAAUCAUAUGCAUAAAGGGAG____UGACCCGU_________UAAUUAUGGUGAAAGAGUGAAGGAAAUCCUAAAUAAUGCAAAUCAAG .((....))..................(((((.......)))))...................................((((..........(((....)))...........)))).. ( -5.44 = -6.44 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:39 2006