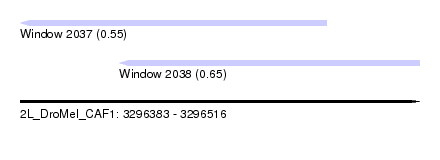
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,296,383 – 3,296,516 |
| Length | 133 |
| Max. P | 0.652921 |

| Location | 3,296,383 – 3,296,485 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.33 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
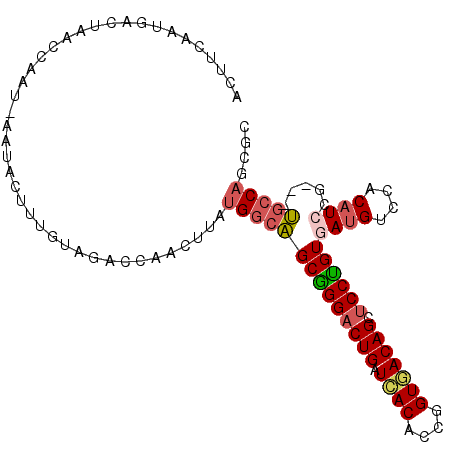
>2L_DroMel_CAF1 3296383 102 - 22407834 ACCAACUUAUGGCAGCGGGACUGAUCACACCGGUGACAGCACCUGUGAUGUCCACAUCCG---UGCCAGCGCAUUUCAUGCAGGGCAACACCUUCAAGCCACUCA .........((((....((((..((((((..((((....)))))))))))))).......---((((...((((...))))..))))..........)))).... ( -31.10) >DroPse_CAF1 6054 105 - 1 ACCAACUAAUGGCAGCAGGACUGAUCACACCAGUCACAGCUCCUGUAAUGUCGACCUCUGGCCUUCCCGCGCACUACCACCAGUCGAGCAGCUUUAAGCCCCUGA ..........(((.(((((((((...((....))..))).))))))..(((((((...(((...............)))...))))).)).......)))..... ( -22.76) >DroSec_CAF1 5144 102 - 1 ACCAUCUUAUGGCAGCGGGACUGAUCACACCGGUGACAGCUCCUGUAAUGUCCACAUCAG---UGACAGCGCAUUUCAUGCAGUGCAACACCUUCAAGCCACUCA .........((((.(((((((((.((((....))))))).))))))..((((.((....)---)))))..(((((......)))))...........)))).... ( -29.60) >DroEre_CAF1 13595 102 - 1 ACCAACUUAUGGCGGCGGGACUGAUCACACCCGUGACAGCUCCCGUGAUGUCCACAUCCG---CGCCAGGGCAUUUCCUGCAGGGCAGCACCUUUAAGCCACUCA .........((((.(((((((((.((((....))))))).))))))((((....)))).(---((((...(((.....)))..))).))........)))).... ( -37.60) >DroYak_CAF1 5248 102 - 1 ACCAACUUAUGGCAGCGGGACUGAUGACACCCGUCACAGCUCCCGUGAUGUCCGCAUCCG---UGCCACCGCAUUUCACGCAGGGCAGCACAUUCAAGCCACUCA .........((((.(((((((((.((((....))))))).))))))((((....)))).(---(((..((((.......)).))...))))......)))).... ( -36.40) >DroPer_CAF1 6027 105 - 1 ACCAGCUAAUGGCAGCAGGACUGAUCACACCAGUCACAGCUCCUGUAAUGUCGACCUCUGGCCUUCCCGCGCACUACCACCAGUCGAGCAGCUUUAAGCCCCUGA ....(((...(((.(((((((((...((....))..))).))))))..(((((((...(((...............)))...))))).)))))...)))...... ( -24.56) >consensus ACCAACUUAUGGCAGCGGGACUGAUCACACCAGUCACAGCUCCUGUAAUGUCCACAUCCG___UGCCAGCGCAUUUCAUGCAGGGCAGCACCUUCAAGCCACUCA ..........(((.(((((((((.((((....))))))).))))))...................................(((......)))....)))..... (-19.44 = -19.33 + -0.11)



| Location | 3,296,416 – 3,296,516 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -20.42 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3296416 100 - 22407834 GCUUCAGUGGCUAACCAAA-AAUUCUUUGUAGACCAACUUAUGGCAGCGGGACUGAUCACACCGGUGACAGCACCUGUGAUGUCCACAUCCG---UGCCAGCGC ((.....((((((..((((-.....))))))).))).....(((((.((((..((..(((((.((((....)))).))).))..))..))))---))))))).. ( -29.90) >DroPse_CAF1 6087 104 - 1 UCUUCACUCACUCUCUGCUCUGUAUGUUCCAGACCAACUAAUGGCAGCAGGACUGAUCACACCAGUCACAGCUCCUGUAAUGUCGACCUCUGGCCUUCCCGCGC ................((((((.......))))(((.....((((((((((((((...((....))..))).))))))..))))).....))).......)).. ( -20.00) >DroSec_CAF1 5177 100 - 1 ACUUCAAUGGCUAAUCAAU-AAUACUUUGUAGACCAUCUUAUGGCAGCGGGACUGAUCACACCGGUGACAGCUCCUGUAAUGUCCACAUCAG---UGACAGCGC ......(((((((..(((.-......)))))).))))......((.(((((((((.((((....))))))).))))))..((((.((....)---))))))).. ( -26.10) >DroSim_CAF1 5193 100 - 1 ACUUCAAUGGCUAACCAAU-AAUACUUUGUAGACCAACUUAUGGCAGCGGGACUGAUCACACCGGUGACAGCUCCUGUGAUGUCCACAUCCG---UGCCAGCGC .......((((((..(((.-......)))))).))).....((((((((((((((.((((....))))))).))))))((((....))))..---))))).... ( -29.70) >DroEre_CAF1 13628 100 - 1 AGUACAAUGACUAACCAAU-UAUACUUGGUAGACCAACUUAUGGCGGCGGGACUGAUCACACCCGUGACAGCUCCCGUGAUGUCCACAUCCG---CGCCAGGGC ..............((...-.....((((....))))....((((((((((((((.((((....))))))).))))))((((....))))..---))))))).. ( -34.60) >DroYak_CAF1 5281 100 - 1 AAAUCAAUGACUAACCAAU-UCUACUUUGUAGACCAACUUAUGGCAGCGGGACUGAUGACACCCGUCACAGCUCCCGUGAUGUCCGCAUCCG---UGCCACCGC ...................-(((((...)))))........((((((((((((((.((((....))))))).))))))((((....))))..---))))).... ( -30.60) >consensus ACUUCAAUGACUAACCAAU_AAUACUUUGUAGACCAACUUAUGGCAGCGGGACUGAUCACACCGGUGACAGCUCCUGUGAUGUCCACAUCCG___UGCCAGCGC .........................................((((((((((((((.((((....))))))).))))))((((....)))).....))))).... (-20.42 = -20.90 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:37 2006