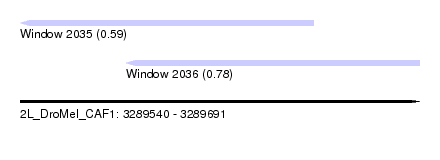
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,289,540 – 3,289,691 |
| Length | 151 |
| Max. P | 0.781331 |

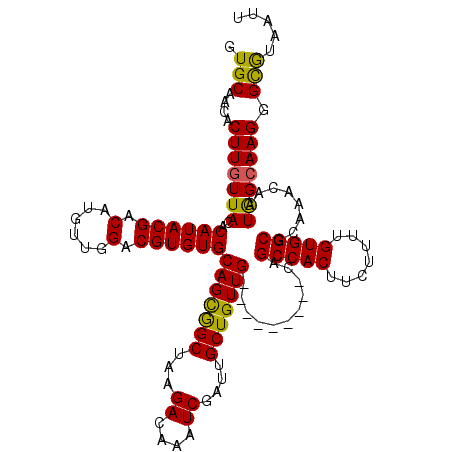
| Location | 3,289,540 – 3,289,651 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.69 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -21.85 |
| Energy contribution | -21.57 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.590931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3289540 111 - 22407834 AGCGGCUAAGACAAAUCGAUUGCUGUUG---------CAGCCACUUCUUUUGUGGCGCAAACAAUGGCAAGGGCGUAAUUUAUUGAACACUUUGUUGCACUUGCCACCGCCGAGCAAUUC ..((((............((((...(((---------(.(((((.......))))))))).))))((((((.(((....................))).))))))...))))........ ( -33.55) >DroSec_CAF1 13441 90 - 1 AGUGGCUAAGACAAAUCGAUUGCUGUUG---------CAGCCACUUCUUUUGUGGCGCAAACAAUGGCAAGGGCGUAAUUUAUUGAACACUUUGUUGCA--------------------- .(..((.(((.........(((((((((---------..(((((.......))))).....)))))))))(..((........))..).))).))..).--------------------- ( -25.60) >DroSim_CAF1 13921 111 - 1 AGUGGCUAAGACAAAUCGAUUGCUGUUG---------CAGCCACUUCUUUUGUGGCGCAAACAAUGGCAAGGGCGUAAUUUAUUGAACACUUUGUUGCACUUGCCACCGCCGAGCAAUUC .................(((((((.(((---------(.(((((.......)))))))))....(((((((.(((....................))).)))))))......))))))). ( -31.85) >DroYak_CAF1 13640 111 - 1 AGCGGCUAAGACAAAUCGAUUGCUGUUG---------CAGCCACUUCUUUUGUGGCGCAAACAAUGGCAAGGGCGUAAUUUAUUGAACACUUUGUUGCACUUGCCACCGCCGAGUAAUUC ..((((............((((...(((---------(.(((((.......))))))))).))))((((((.(((....................))).))))))...))))........ ( -33.55) >DroAna_CAF1 28906 118 - 1 AGCAGCUAAGACAAAUCGAUGGCUGUUGCCGUUGCAGCAGCCACUUCUUUUGUGGCACAAAAAAUAGAAAGGGUAAAAUUUAUUGAACACUUUGUUGCACUCGCCGUCACUUUUAAAC-- .(((((.(((.....((((((((((((((....)))))))))(((..((((((..........))))))..))).......)))))...))).)))))....................-- ( -31.90) >consensus AGCGGCUAAGACAAAUCGAUUGCUGUUG_________CAGCCACUUCUUUUGUGGCGCAAACAAUGGCAAGGGCGUAAUUUAUUGAACACUUUGUUGCACUUGCCACCGCCGAGCAAUUC .(((((.(((.........(((((((((...........(((((.......))))).....)))))))))(..((........))..).))).)))))...................... (-21.85 = -21.57 + -0.28)

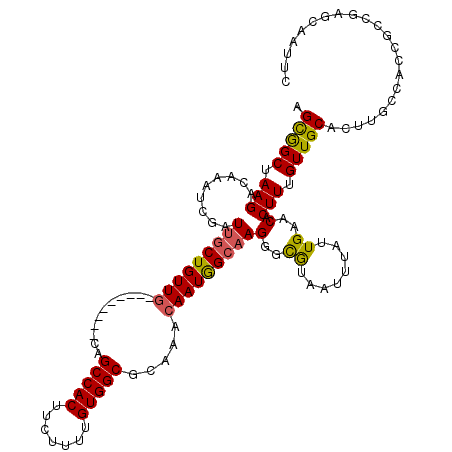
| Location | 3,289,580 – 3,289,691 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.80 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -27.96 |
| Energy contribution | -27.28 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3289580 111 - 22407834 GUGCAACACUUGUUACACAUACGACAUGUUGGACGUGUGCAGCGGCUAAGACAAAUCGAUUGCUGUUG---------CAGCCACUUCUUUUGUGGCGCAAACAAUGGCAAGGGCGUAAUU ..((....((((((((((...(((....)))...)))(((((((((...((....))....)))))))---------))(((((.......)))))........))))))).))...... ( -34.60) >DroSec_CAF1 13460 111 - 1 GUGCAACACUUGUUACACAUACGACAUGUUGGACGUGUGCAGUGGCUAAGACAAAUCGAUUGCUGUUG---------CAGCCACUUCUUUUGUGGCGCAAACAAUGGCAAGGGCGUAAUU ..((....((((((((((...(((....)))...)))(((((..((...((....))....))..)))---------))(((((.......)))))........))))))).))...... ( -31.70) >DroSim_CAF1 13961 111 - 1 GUGCAACACUUGUUACACAUACGACAUGUUGGACGUGUGCAGUGGCUAAGACAAAUCGAUUGCUGUUG---------CAGCCACUUCUUUUGUGGCGCAAACAAUGGCAAGGGCGUAAUU ..((....((((((((((...(((....)))...)))(((((..((...((....))....))..)))---------))(((((.......)))))........))))))).))...... ( -31.70) >DroYak_CAF1 13680 111 - 1 GUGCAACGCUUGUUACACAUACGACAUGUUGGACGUGUGCAGCGGCUAAGACAAAUCGAUUGCUGUUG---------CAGCCACUUCUUUUGUGGCGCAAACAAUGGCAAGGGCGUAAUU .....(((((((((((((...(((....)))...)))(((((((((...((....))....)))))))---------))(((((.......)))))........)))))..))))).... ( -34.40) >DroAna_CAF1 28944 120 - 1 GUGCUGCACUUGUUACACAUACGACAUGUGGGACGUGUGCAGCAGCUAAGACAAAUCGAUGGCUGUUGCCGUUGCAGCAGCCACUUCUUUUGUGGCACAAAAAAUAGAAAGGGUAAAAUU .((((((((.(((..(((((.....)))))..))).))))))))(((...(((((..((((((((((((....)))))))))).))..))))))))........................ ( -46.50) >consensus GUGCAACACUUGUUACACAUACGACAUGUUGGACGUGUGCAGCGGCUAAGACAAAUCGAUUGCUGUUG_________CAGCCACUUCUUUUGUGGCGCAAACAAUGGCAAGGGCGUAAUU .(((....(((((((..((((((.(......).))))))(((((((...((....))....)))))))...........(((((.......)))))........))))))).)))..... (-27.96 = -27.28 + -0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:35 2006