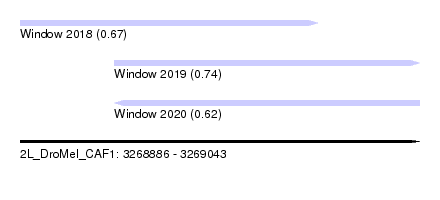
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,268,886 – 3,269,043 |
| Length | 157 |
| Max. P | 0.735519 |

| Location | 3,268,886 – 3,269,003 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.55 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -22.86 |
| Energy contribution | -23.61 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3268886 117 + 22407834 CCGAGUA---AUACGCGUUAGCUUAACAGGGAAUUUAGGAGCAUUUUAUUUAAUUUUAAAUUCCAUCAAACUUUUGCGCUGUGCCAAGUGCAGCAAUGGCGCAAUUCAACUAUCUAAAUU ..((((.---....((((...........(((((((((((.............)))))))))))...........))))(((((((..........)))))))))))............. ( -26.37) >DroSec_CAF1 95710 117 + 1 CCGAGUA---AUGCGCGUUAGCCUAACCGGGAAUUUAGGAGCAUUUUAUUUAAUUUUAAAUUCCAUCAAACUUUUGCGCUGUGCCAAGUGCAGCAAUGGCGCAAUUCAACUAUCUAAAUU ..((((.---.(((((((...........(((((((((((.............)))))))))))...........))((((..(...)..))))....)))))))))............. ( -29.57) >DroSim_CAF1 99161 117 + 1 CCGUAAA---AUGCGCGUUAGCUUAACCGGGAAUUUAGGAGCAUUUUAUUUAAUUUUAAAUUCCAUCAAACUUUUGCGCUGUGCCAAGUGCAGCAAUGGCGCAAUUCAACUAUCUAAAUU .......---.(((((((...........(((((((((((.............)))))))))))...........))((((..(...)..))))....)))))................. ( -27.57) >DroYak_CAF1 92240 120 + 1 ACGAUUACUACUACGCGUAAGCUUAACUGGAAAGUUAGGAGCUUUUAAUUUAAUUUUAAAUUCCAUCAAACUUUUGCGCUGUGCCAAGUGCAGCAAUGGCGCAAUUCAACUAUCUAAAUU ..............((....)).....((((.((((.((((..(((((.......))))))))).........(((((((((((........)).)))))))))...)))).)))).... ( -25.20) >consensus CCGAGUA___AUACGCGUUAGCUUAACCGGGAAUUUAGGAGCAUUUUAUUUAAUUUUAAAUUCCAUCAAACUUUUGCGCUGUGCCAAGUGCAGCAAUGGCGCAAUUCAACUAUCUAAAUU ..............((((((((.......(((((((((((.............)))))))))))...........))((((..(...)..))))..)))))).................. (-22.86 = -23.61 + 0.75)

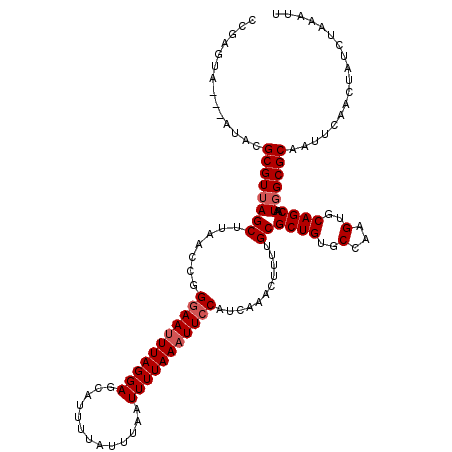

| Location | 3,268,923 – 3,269,043 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -23.80 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3268923 120 + 22407834 GCAUUUUAUUUAAUUUUAAAUUCCAUCAAACUUUUGCGCUGUGCCAAGUGCAGCAAUGGCGCAAUUCAACUAUCUAAAUUGCAAAAACCAGACACAUGAUGGAAAAACGUUAAUGGCUCC ((..((((........))))((((((((....((((.((((..(...)..))))......((((((..........))))))......))))....))))))))...........))... ( -24.80) >DroSec_CAF1 95747 116 + 1 GCAUUUUAUUUAAUUUUAAAUUCCAUCAAACUUUUGCGCUGUGCCAAGUGCAGCAAUGGCGCAAUUCAACUAUCUAAAUUGCAAAA----GGCACAUGAUGGAAAAACGUUAAUGGCUCC ((..((((........))))((((((((..(((((((((((..(...)..))))..(((......)))............))))))----).....))))))))...........))... ( -27.40) >DroSim_CAF1 99198 120 + 1 GCAUUUUAUUUAAUUUUAAAUUCCAUCAAACUUUUGCGCUGUGCCAAGUGCAGCAAUGGCGCAAUUCAACUAUCUAAAUUGCAAAAACCAGGCACAUGAUGGAAAAACGUUAAUGGCUCC ((..((((........))))((((((((......((((((......))))))((..(((.((((((..........)))))).....))).))...))))))))...........))... ( -25.80) >DroYak_CAF1 92280 120 + 1 GCUUUUAAUUUAAUUUUAAAUUCCAUCAAACUUUUGCGCUGUGCCAAGUGCAGCAAUGGCGCAAUUCAACUAUCUAAAUUGCAAAAACCAGGCACAUGAUGGAAAAACGUUAAUGGCUCC (((.((((((((....))).((((((((......((((((......))))))((..(((.((((((..........)))))).....))).))...))))))))....))))).)))... ( -27.40) >consensus GCAUUUUAUUUAAUUUUAAAUUCCAUCAAACUUUUGCGCUGUGCCAAGUGCAGCAAUGGCGCAAUUCAACUAUCUAAAUUGCAAAAACCAGGCACAUGAUGGAAAAACGUUAAUGGCUCC ....................((((((((.........((((..(...)..))))......((((((..........))))))..............))))))))................ (-23.80 = -23.80 + 0.00)

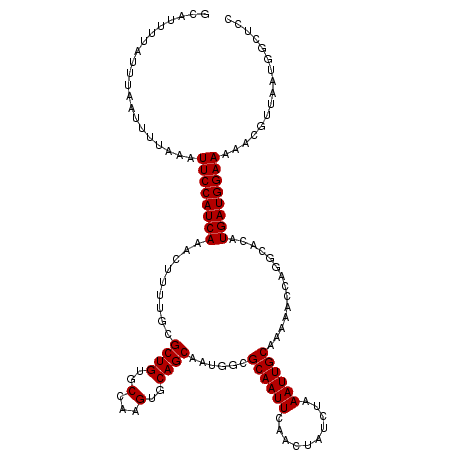
| Location | 3,268,923 – 3,269,043 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -28.09 |
| Energy contribution | -27.90 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3268923 120 - 22407834 GGAGCCAUUAACGUUUUUCCAUCAUGUGUCUGGUUUUUGCAAUUUAGAUAGUUGAAUUGCGCCAUUGCUGCACUUGGCACAGCGCAAAAGUUUGAUGGAAUUUAAAAUUAAAUAAAAUGC .......((((..((.((((((((((((.(((......(((((((((....)))))))))((((..........)))).)))))))......))))))))...))..))))......... ( -29.60) >DroSec_CAF1 95747 116 - 1 GGAGCCAUUAACGUUUUUCCAUCAUGUGCC----UUUUGCAAUUUAGAUAGUUGAAUUGCGCCAUUGCUGCACUUGGCACAGCGCAAAAGUUUGAUGGAAUUUAAAAUUAAAUAAAAUGC .......((((..((.(((((((((((((.----....(((((((((....)))))))))((((..........))))...)))))......))))))))...))..))))......... ( -28.40) >DroSim_CAF1 99198 120 - 1 GGAGCCAUUAACGUUUUUCCAUCAUGUGCCUGGUUUUUGCAAUUUAGAUAGUUGAAUUGCGCCAUUGCUGCACUUGGCACAGCGCAAAAGUUUGAUGGAAUUUAAAAUUAAAUAAAAUGC .......((((..((.((((((((..((((((......(((((((((....)))))))))((((..........)))).))).)))......))))))))...))..))))......... ( -30.50) >DroYak_CAF1 92280 120 - 1 GGAGCCAUUAACGUUUUUCCAUCAUGUGCCUGGUUUUUGCAAUUUAGAUAGUUGAAUUGCGCCAUUGCUGCACUUGGCACAGCGCAAAAGUUUGAUGGAAUUUAAAAUUAAAUUAAAAGC .......((((..((.((((((((..((((((......(((((((((....)))))))))((((..........)))).))).)))......))))))))...))..))))......... ( -30.50) >consensus GGAGCCAUUAACGUUUUUCCAUCAUGUGCCUGGUUUUUGCAAUUUAGAUAGUUGAAUUGCGCCAUUGCUGCACUUGGCACAGCGCAAAAGUUUGAUGGAAUUUAAAAUUAAAUAAAAUGC .......((((..((.(((((((((((((.........(((((((((....)))))))))((((..........))))...)))))......))))))))...))..))))......... (-28.09 = -27.90 + -0.19)

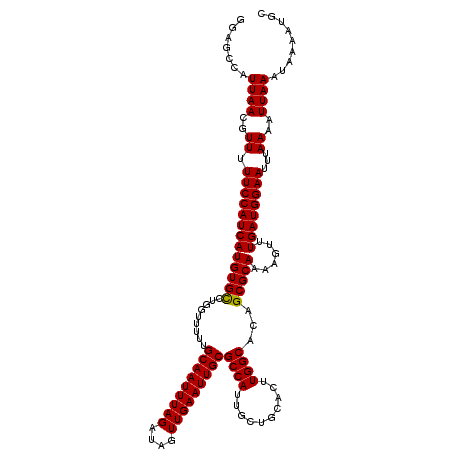
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:19 2006