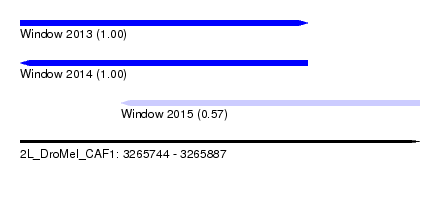
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,265,744 – 3,265,887 |
| Length | 143 |
| Max. P | 0.999417 |

| Location | 3,265,744 – 3,265,847 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.32 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -24.30 |
| Energy contribution | -27.26 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.74 |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.999417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3265744 103 + 22407834 --AAUGU-AUCGCU-AAGGGAAAUUUUUCCGGGGAAAAGGUCGAAAGACCUCUCCCAACCGUGCAACACGUUCGCUGCAACAUGUUGCAUGGGGACACAUCCUUUUA --.....-......-((((((......(((.((((..(((((....))))).))))..((((((((((.(((......))).)))))))))))))....)))))).. ( -40.90) >DroSec_CAF1 92406 106 + 1 AUAAUGUAAUCGCU-AAGGGAAAUUUUUCCGGGGAAAAGGUCGAAAGACCUCUCCCAACCGUGCAACACGUUCGCUGCAACAUGUUGCAUGGGGACACAUCGUUUUA ...((((.......-...((((....)))).((((..(((((....))))).))))..((((((((((.(((......))).))))))))))....))))....... ( -37.70) >DroSim_CAF1 95487 106 + 1 AUAAUGUAAUCGCU-AAGGGAAAUUUUUCCGGGGAAAAGGUCGAAAGACCUCUCCCAACCGUGCAACNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ....((((......-...((((....)))).((((..(((((....))))).)))).....)))).......................................... ( -21.60) >DroEre_CAF1 89123 102 + 1 ---AUGU-UUCGCA-AGGGGAAAUUUUUCCGAGGAAAAGGUUGAAAGACCUAUGCCAACCGUGCAACACGUUCGCUGCAACAUGUUGCAUGGGGACACAACCUUUUA ---...(-..(...-.)..).......(((..((...(((((....)))))...))..((((((((((.(((......))).)))))))))))))............ ( -33.70) >DroYak_CAF1 88791 103 + 1 ---AUGU-UUCGCAGAAGGGAAAUUUUUCCGAGGAAAAGAUUGAAAGACCUCUCCCAACCGUGCAACACGUUCGCUGCAACAUGUUGCAUGGGGACACAGUCUUUUA ---..((-(((.(....).)))))(((((....)))))...((((((((...((((...(((((((((.(((......))).)))))))))))))....)))))))) ( -31.10) >consensus __AAUGU_AUCGCU_AAGGGAAAUUUUUCCGGGGAAAAGGUCGAAAGACCUCUCCCAACCGUGCAACACGUUCGCUGCAACAUGUUGCAUGGGGACACAUCCUUUUA .................((((...(((((....)))))((((....))))..))))..((((((((((.(((......))).))))))))))............... (-24.30 = -27.26 + 2.96)

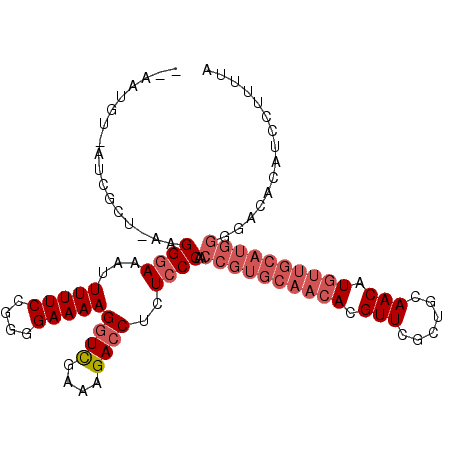
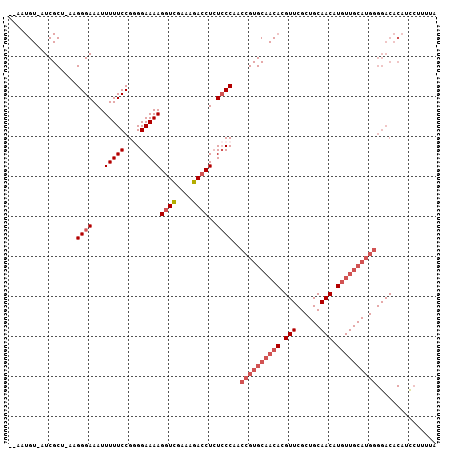
| Location | 3,265,744 – 3,265,847 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.32 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -23.85 |
| Energy contribution | -26.34 |
| Covariance contribution | 2.49 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.999109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3265744 103 - 22407834 UAAAAGGAUGUGUCCCCAUGCAACAUGUUGCAGCGAACGUGUUGCACGGUUGGGAGAGGUCUUUCGACCUUUUCCCCGGAAAAAUUUCCCUU-AGCGAU-ACAUU-- ......((((((((.((.(((((((((((......))))))))))).))..((((((((((....))))))))))..((((....))))...-...)))-)))))-- ( -41.60) >DroSec_CAF1 92406 106 - 1 UAAAACGAUGUGUCCCCAUGCAACAUGUUGCAGCGAACGUGUUGCACGGUUGGGAGAGGUCUUUCGACCUUUUCCCCGGAAAAAUUUCCCUU-AGCGAUUACAUUAU ......((((((((.((.(((((((((((......))))))))))).))..((((((((((....))))))))))..((((....))))...-...)).)))))).. ( -38.40) >DroSim_CAF1 95487 106 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGUUGCACGGUUGGGAGAGGUCUUUCGACCUUUUCCCCGGAAAAAUUUCCCUU-AGCGAUUACAUUAU ........................................(((((..((..((((((((((....))))))))))))((((....))))...-.)))))........ ( -23.10) >DroEre_CAF1 89123 102 - 1 UAAAAGGUUGUGUCCCCAUGCAACAUGUUGCAGCGAACGUGUUGCACGGUUGGCAUAGGUCUUUCAACCUUUUCCUCGGAAAAAUUUCCCCU-UGCGAA-ACAU--- .(((((((((((((.((.(((((((((((......))))))))))).))..)))).........)))))))))....((((....))))...-......-....--- ( -30.30) >DroYak_CAF1 88791 103 - 1 UAAAAGACUGUGUCCCCAUGCAACAUGUUGCAGCGAACGUGUUGCACGGUUGGGAGAGGUCUUUCAAUCUUUUCCUCGGAAAAAUUUCCCUUCUGCGAA-ACAU--- ..(((((((...(((((.(((((((((((......))))))))))).))..)))...))))))).....(((((....))))).((((.(....).)))-)...--- ( -30.70) >consensus UAAAAGGAUGUGUCCCCAUGCAACAUGUUGCAGCGAACGUGUUGCACGGUUGGGAGAGGUCUUUCGACCUUUUCCCCGGAAAAAUUUCCCUU_AGCGAU_ACAUU__ ...............((.(((((((((((......))))))))))).))..((((((((((....))))))))))..((((....)))).................. (-23.85 = -26.34 + 2.49)

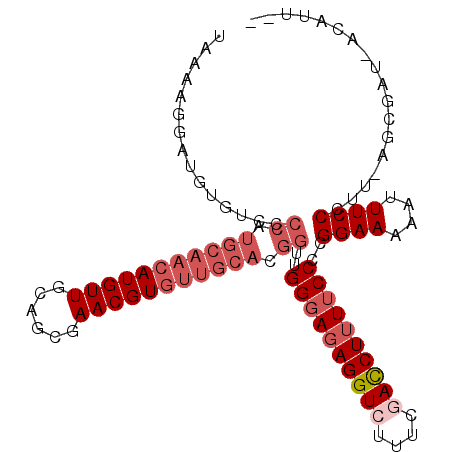
| Location | 3,265,780 – 3,265,887 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 93.93 |
| Mean single sequence MFE | -31.81 |
| Consensus MFE | -25.73 |
| Energy contribution | -25.72 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3265780 107 - 22407834 CCGUGGAUCUGUGACAAGUUCCACACAAUGCGUAAACAAAUAAAAGGAUGUGUCCCCAUGCAACAUGUUGCAGCGAACGUGUUGCACGGUUGGGAGAGGUCUUUCGA ..(((((.((......)).)))))...................((((((.(.(((((.(((((((((((......))))))))))).)...)))).).))))))... ( -35.90) >DroSec_CAF1 92445 103 - 1 ---UGGAUCUGUGACAAG-UCCACACAAUGCGUAAACAAAUAAAACGAUGUGUCCCCAUGCAACAUGUUGCAGCGAACGUGUUGCACGGUUGGGAGAGGUCUUUCGA ---.((((((.(..(((.-...(((((...(((...........))).)))))..((.(((((((((((......))))))))))).)))))..).))))))..... ( -30.90) >DroEre_CAF1 89158 107 - 1 CAGUGAAUCUGUGACAAGUUCCACACAAUGCGUAAACAAAUAAAAGGUUGUGUCCCCAUGCAACAUGUUGCAGCGAACGUGUUGCACGGUUGGCAUAGGUCUUUCAA ...((((((((((.(((.....(((((((.................)))))))..((.(((((((((((......))))))))))).))))).)))))))...))). ( -29.93) >DroYak_CAF1 88827 107 - 1 CAGUGAAUCUGUGACAAGUUCCACACAAUGCGUAAACAAAUAAAAGACUGUGUCCCCAUGCAACAUGUUGCAGCGAACGUGUUGCACGGUUGGGAGAGGUCUUUCAA ..(((...((......))...)))..................(((((((...(((((.(((((((((((......))))))))))).))..)))...)))))))... ( -30.50) >consensus CAGUGAAUCUGUGACAAGUUCCACACAAUGCGUAAACAAAUAAAAGGAUGUGUCCCCAUGCAACAUGUUGCAGCGAACGUGUUGCACGGUUGGGAGAGGUCUUUCAA ...((((.....(((...(((((((((.....................)))))..((.(((((((((((......))))))))))).))..))))...))).)))). (-25.73 = -25.72 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:14 2006