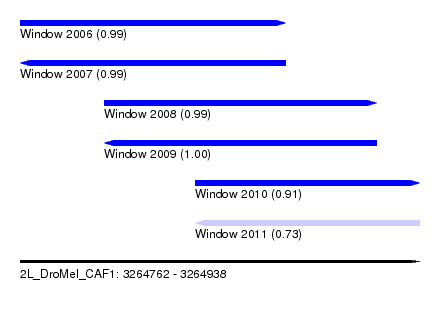
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,264,762 – 3,264,938 |
| Length | 176 |
| Max. P | 0.998752 |

| Location | 3,264,762 – 3,264,879 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -22.42 |
| Energy contribution | -24.02 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3264762 117 + 22407834 AAAAUUUCUCCCCCAUUUUCCUUUGAUUUUAUCGGCGUGCAAGCAACUUGUUUGCUUUCACUUUGCUUCGCUUUUGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUU ..........(((((.......(((((((..((((((((.((((((.....)))))).))).((((...((....)).))))........))))).....))))))).))))).... ( -28.30) >DroSec_CAF1 91404 117 + 1 AAAAUUUCUCCCCCAUUUUCGUUCGAUUUUAUCGGCGUGCAAGCAACUUGUUUGCUUUCACUUUGCUUCGCUUUUGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUU ..........(((((.....(((.(((((..((((((((.((((((.....)))))).))).((((...((....)).))))........))))).....))))))))))))).... ( -26.20) >DroSim_CAF1 94469 117 + 1 AAAAUUUCUCCCCCAUUUUCGUUUGAUUUUAUCGGCGUGCAAGCAACUUGUUUGCUUUCACUUUGCUUCGCUUUUGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUU ..........(((((.......(((((((..((((((((.((((((.....)))))).))).((((...((....)).))))........))))).....))))))).))))).... ( -28.30) >DroEre_CAF1 88280 115 + 1 AAAAAUUUCCCC--CUUUUCGUUUGAUUUUAUCGGCGUGCAAGCAACUUGUUUGCUUUCACUUUGCUUCGCAUUCGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGAGGGUUU ..........((--(((.....(((((((.((((((((((((((((..((........))..)))))).)))..)))).(((((.....))))))))...)))))))..)))))... ( -28.90) >DroYak_CAF1 87785 113 + 1 AAAAUUUUUCCCCCAUUUUCGUUUGAUUUUAUCGGCGUGC----AACUUGUUUGCUUUCACUUUGCUUCGCCUUUGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUU ..........(((((.......(((((((.(((((((.((----((..((........))..))))..)))).......(((((.....))))))))...))))))).))))).... ( -29.30) >consensus AAAAUUUCUCCCCCAUUUUCGUUUGAUUUUAUCGGCGUGCAAGCAACUUGUUUGCUUUCACUUUGCUUCGCUUUUGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUU ..........(((((.......(((((((..((((((((.((((((.....)))))).))).((((...((....)).))))........))))).....))))))).))))).... (-22.42 = -24.02 + 1.60)


| Location | 3,264,762 – 3,264,879 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.44 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3264762 117 - 22407834 AAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACAAAAGCGAAGCAAAGUGAAAGCAAACAAGUUGCUUGCACGCCGAUAAAAUCAAAGGAAAAUGGGGGAGAAAUUUU ....(((((.(((((((...(((..((.........(((..(......)..)))..(((.((((((.....)))))).))))).))).))))))).......))))).......... ( -23.20) >DroSec_CAF1 91404 117 - 1 AAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACAAAAGCGAAGCAAAGUGAAAGCAAACAAGUUGCUUGCACGCCGAUAAAAUCGAACGAAAAUGGGGGAGAAAUUUU ....(((((.(((((((...(((..((.........(((..(......)..)))..(((.((((((.....)))))).))))).))).))))))).......))))).......... ( -22.90) >DroSim_CAF1 94469 117 - 1 AAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACAAAAGCGAAGCAAAGUGAAAGCAAACAAGUUGCUUGCACGCCGAUAAAAUCAAACGAAAAUGGGGGAGAAAUUUU ....(((((.(((((((...(((..((.........(((..(......)..)))..(((.((((((.....)))))).))))).))).))))))).......))))).......... ( -23.20) >DroEre_CAF1 88280 115 - 1 AAACCCUCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACGAAUGCGAAGCAAAGUGAAAGCAAACAAGUUGCUUGCACGCCGAUAAAAUCAAACGAAAAG--GGGGAAAUUUUU ...(((((..(((((((...(((..((.........(((..((....))..)))..(((.((((((.....)))))).))))).))).))))))).......)--))))........ ( -24.30) >DroYak_CAF1 87785 113 - 1 AAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACAAAGGCGAAGCAAAGUGAAAGCAAACAAGUU----GCACGCCGAUAAAAUCAAACGAAAAUGGGGGAAAAAUUUU ....(((((.(((((((...((((((.........))).......((((..((((..((........))..))----)).))))))).))))))).......))))).......... ( -23.30) >consensus AAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACAAAAGCGAAGCAAAGUGAAAGCAAACAAGUUGCUUGCACGCCGAUAAAAUCAAACGAAAAUGGGGGAGAAAUUUU ....((((..(((((((...(((..((.........(((..(......)..)))..(((.((((((.....)))))).))))).))).))))))).........))))......... (-18.76 = -19.44 + 0.68)



| Location | 3,264,799 – 3,264,919 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -23.90 |
| Energy contribution | -25.30 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3264799 120 + 22407834 UGCAAGCAACUUGUUUGCUUUCACUUUGCUUCGCUUUUGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUUAUUUUGCCCUAUCCUCAGCCAUUGCCAAUUUCAAUUGCCG .((((((((..((........))..)))))).)).......((((((..(((((((((.............((((((........)))))).......)))..))))))...)))))).. ( -29.45) >DroSec_CAF1 91441 120 + 1 UGCAAGCAACUUGUUUGCUUUCACUUUGCUUCGCUUUUGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUUAUUUUGCCCUAGCCUCAGCCAUUGCCAAUUUCAAUUGCCG .((((((((..((........))..)))))).)).......((((((..((((((..(((.....)))..(((((((........)))))))...........))))))...)))))).. ( -29.90) >DroSim_CAF1 94506 119 + 1 UGCAAGCAACUUGUUUGCUUUCACUUUGCUUCGCUUUUGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUUAUUCUGCCCUAGCCUCAGCCAUUGCCAAUUUCAA-UGCCG .(((((((((..((..((.........))...))....)))))........(((((((((.....)))..(((((((........)))))))...)))))))))).........-..... ( -29.60) >DroEre_CAF1 88315 120 + 1 UGCAAGCAACUUGUUUGCUUUCACUUUGCUUCGCAUUCGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGAGGGUUUGUUUUGCCAUAGCCUUUGCUAUUGCCAAUUUCAAUUGCUA (((((((((..((........))..)))))).)))......((((((..((((((.((....((((((.......)))))).....))(((((....))))).))))))...)))))).. ( -30.50) >DroYak_CAF1 87822 116 + 1 UGC----AACUUGUUUGCUUUCACUUUGCUUCGCCUUUGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUUAUUUUGCCCUAGCCUUUGCUAUUGCCAAUUUCAAUUGCCG .((----((..((........))..))))............((((((..((((((..(((.....)))..(((((((........)))))))...........))))))...)))))).. ( -25.20) >consensus UGCAAGCAACUUGUUUGCUUUCACUUUGCUUCGCUUUUGUUGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUUAUUUUGCCCUAGCCUCAGCCAUUGCCAAUUUCAAUUGCCG .((((((((..((........))..)))))).)).......((((((..((((((..(((.....)))...((((((........))))))............))))))...)))))).. (-23.90 = -25.30 + 1.40)


| Location | 3,264,799 – 3,264,919 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -22.58 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3264799 120 - 22407834 CGGCAAUUGAAAUUGGCAAUGGCUGAGGAUAGGGCAAAAUAAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACAAAAGCGAAGCAAAGUGAAAGCAAACAAGUUGCUUGCA .(.((((....)))).)..((((((......(((...........)))....(((.....))))))))).................((.((((((..((........))..)))))))). ( -25.20) >DroSec_CAF1 91441 120 - 1 CGGCAAUUGAAAUUGGCAAUGGCUGAGGCUAGGGCAAAAUAAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACAAAAGCGAAGCAAAGUGAAAGCAAACAAGUUGCUUGCA ..(((.((...((((((..((((....))))(((...........))).................))))))..)).))).......((.((((((..((........))..)))))))). ( -28.10) >DroSim_CAF1 94506 119 - 1 CGGCA-UUGAAAUUGGCAAUGGCUGAGGCUAGGGCAGAAUAAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACAAAAGCGAAGCAAAGUGAAAGCAAACAAGUUGCUUGCA ..(((-.....((((((..((((....))))(((...........))).................)))))).....))).......((.((((((..((........))..)))))))). ( -27.60) >DroEre_CAF1 88315 120 - 1 UAGCAAUUGAAAUUGGCAAUAGCAAAGGCUAUGGCAAAACAAACCCUCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACGAAUGCGAAGCAAAGUGAAAGCAAACAAGUUGCUUGCA ..(((.((...((((((.(((((....))))).......(((........)))............))))))..)).)))......(((.((((((..((........))..))))))))) ( -25.90) >DroYak_CAF1 87822 116 - 1 CGGCAAUUGAAAUUGGCAAUAGCAAAGGCUAGGGCAAAAUAAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACAAAGGCGAAGCAAAGUGAAAGCAAACAAGUU----GCA ..(((.((...((((((..((((....))))(((...........))).................))))))..)).)))............((((..((........))..))----)). ( -23.30) >consensus CGGCAAUUGAAAUUGGCAAUGGCUGAGGCUAGGGCAAAAUAAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCAACAAAAGCGAAGCAAAGUGAAAGCAAACAAGUUGCUUGCA ..(((.((...((((((..((((....))))(((...........))).................))))))..)).))).......((.((((((..((........))..)))))))). (-22.58 = -23.38 + 0.80)


| Location | 3,264,839 – 3,264,938 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.65 |
| Mean single sequence MFE | -31.91 |
| Consensus MFE | -20.89 |
| Energy contribution | -22.58 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3264839 99 + 22407834 UGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUUAUUUUGCCCUAUCCUCAGCCAUUGCCAAUUUCAAUUGCCGUUCGCUGUUCAGCGGUUUU--------------------- .((((((..(((((((((.............((((((........)))))).......)))..))))))...))))))...(((((....)))))....--------------------- ( -26.65) >DroSec_CAF1 91481 120 + 1 UGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUUAUUUUGCCCUAGCCUCAGCCAUUGCCAAUUUCAAUUGCCGUUCGCUGUUCAGCGGUUUUCGCUUUGUCAAAGGCAAAAUA ...........(((((((((.....)))..(((((((........)))))))...))))))(((((.....(((..((...(((((....))))).....)).))).....))))).... ( -32.90) >DroSim_CAF1 94546 117 + 1 UGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUUAUUCUGCCCUAGCCUCAGCCAUUGCCAAUUUCAA-UGCCGUUCGCUGUUCAGCGGUU-UCGCU-UGUCAAAGGCAAAAUA ...........(((((((((.....)))..(((((((........)))))))...))))))(((((.....(((-.((...(((((....)))))..-..)))-)).....))))).... ( -33.80) >DroEre_CAF1 88355 120 + 1 UGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGAGGGUUUGUUUUGCCAUAGCCUUUGCUAUUGCCAAUUUCAAUUGCUAUUUGAAGUUCAGCGGUUUUCGCUUUGUCAAAGGCAAAAUA .............(((((((((((((((((((((((..(((....((.(((((....))))).))))))))))))))..)))))))))))))).(((((.(((((....)))))))))). ( -34.30) >DroYak_CAF1 87858 120 + 1 UGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUUAUUUUGCCCUAGCCUUUGCUAUUGCCAAUUUCAAUUGCCGUUUGAAGUUUAGCGGUUUUCGCCUUUUCAAAGGACAAAAG ....(((((.(((((((((((((((((((((((((((........))))((((....))))..........))))))..))))))))))))))(((....)))....))))))))..... ( -31.90) >consensus UGCAGUUUUAUUGGCUGGAUUUUAAAUCAAUUGGGGUUUUAUUUUGCCCUAGCCUCAGCCAUUGCCAAUUUCAAUUGCCGUUCGCUGUUCAGCGGUUUUCGCUUUGUCAAAGGCAAAAUA .((((((..((((((..(((.....)))...((((((........))))))............))))))...))))))...(((((....))))).....(((((....)))))...... (-20.89 = -22.58 + 1.69)


| Location | 3,264,839 – 3,264,938 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.65 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3264839 99 - 22407834 ---------------------AAAACCGCUGAACAGCGAACGGCAAUUGAAAUUGGCAAUGGCUGAGGAUAGGGCAAAAUAAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCA ---------------------.....((((....))))...(.((((....)))).)..((((((......(((...........)))....(((.....)))))))))........... ( -19.80) >DroSec_CAF1 91481 120 - 1 UAUUUUGCCUUUGACAAAGCGAAAACCGCUGAACAGCGAACGGCAAUUGAAAUUGGCAAUGGCUGAGGCUAGGGCAAAAUAAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCA ....(((((((((....((((.....))))......)))).))))).....((((((..((((....))))(((...........))).................))))))......... ( -28.20) >DroSim_CAF1 94546 117 - 1 UAUUUUGCCUUUGACA-AGCGA-AACCGCUGAACAGCGAACGGCA-UUGAAAUUGGCAAUGGCUGAGGCUAGGGCAGAAUAAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCA .....((((((((...-((((.-...))))......)))).))))-.....((((((..((((....))))(((...........))).................))))))......... ( -26.90) >DroEre_CAF1 88355 120 - 1 UAUUUUGCCUUUGACAAAGCGAAAACCGCUGAACUUCAAAUAGCAAUUGAAAUUGGCAAUAGCAAAGGCUAUGGCAAAACAAACCCUCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCA .........(((((...((((.....)))).....)))))..(((.((...((((((.(((((....))))).......(((........)))............))))))..)).))). ( -22.30) >DroYak_CAF1 87858 120 - 1 CUUUUGUCCUUUGAAAAGGCGAAAACCGCUAAACUUCAAACGGCAAUUGAAAUUGGCAAUAGCAAAGGCUAGGGCAAAAUAAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCA (..(((((.((((((..((((.....))))....)))))).)))))..)..((((((..((((....))))(((...........))).................))))))......... ( -27.90) >consensus UAUUUUGCCUUUGACAAAGCGAAAACCGCUGAACAGCGAACGGCAAUUGAAAUUGGCAAUGGCUGAGGCUAGGGCAAAAUAAAACCCCAAUUGAUUUAAAAUCCAGCCAAUAAAACUGCA .................((((.....))))............(((.((...((((((..((((....))))(((...........))).................))))))..)).))). (-16.76 = -17.44 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:10 2006