
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,261,985 – 3,262,081 |
| Length | 96 |
| Max. P | 0.631340 |

| Location | 3,261,985 – 3,262,081 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -24.61 |
| Energy contribution | -26.75 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3261985 96 - 22407834 CAGCGUAAUUGACAUCUGAUUGAAG-AAGCAUCUGGCUGCUUGCCAGGUCC---GC-UACGUGCCACCUGGCUAAUCACACAGCCAGGAACGACAGGUGUA------------------- ...........(((((((.......-.((((((((((.....)))))))..---))-).(((....(((((((........))))))).))).))))))).------------------- ( -36.20) >DroSec_CAF1 88606 111 - 1 CAGCGUAAUUGACAUCUGAUUGAAG-AAGCAUCUGGCUGCUUGCCAGGUCC---GC-UACGUGCCACCUAGCUAAUCACACAGCCAGGCACGACAGGUGUGGA----UAGAGAUGUGGAG ..((((.(((.(((((((.......-.((((((((((.....)))))))..---))-).((((((.....(((........)))..)))))).))))))).))----)....)))).... ( -37.80) >DroSim_CAF1 91720 111 - 1 CAGCGUAAUUGACAUCUGAUUGAAG-AAGCAUCUGGCUGCUUGCCAGGUCC---GC-UACGUGCCACCUGGCUAAUCACACAGCCAGGCACGACAGGUGUGGA----UAGAGAUGUGGAG ..((((.(((.(((((((.......-.((((((((((.....)))))))..---))-).((((((...(((((........))))))))))).))))))).))----)....)))).... ( -43.20) >DroEre_CAF1 85385 111 - 1 CAGCGUAAUUGACAUCUGAUUGAAG-AAGCAUCUGGCUGCUUGCCAGGUCC---GC-UACGUGCCACCUGGCUAAUCACACAGCCCGGCACGACAGGUGUGGA----UGGAGGUGUGUAG ((.(...(((.(((((((.......-.((((((((((.....)))))))..---))-).((((((....((((........)))).)))))).))))))).))----)...).))..... ( -42.50) >DroWil_CAF1 102351 109 - 1 CAACGUAAUUGACGUUUGAUUGAAGGCAGCAUCUGGCUUUGUGCCAACUCUAGGAC-UACGUGCCUUCAAGCUAAUCACAGUGCU----UCAACCUGUGUGA-----U-GAUUUGCGUUG ((((((((....((((...(((((((((.....((((.....))))......(...-..).)))))))))......(((((.(..----....)))))).))-----)-)..)))))))) ( -30.70) >DroYak_CAF1 84881 116 - 1 CAGCGUAAUUGACAUCUGAUUGAAG-AAGCAUCUGGCUGCUUGCCAGGUCC---GCAUACGUGCCACCUGGCUAAUCACACAGCCAGCCACGACAGGUGUAGAUAGAUAGAGAUGUGUAG ..((((.(((.(((((((.......-..(((((((((.....)))))))..---))...((((....((((((........)))))).)))).))))))).....)))....)))).... ( -34.70) >consensus CAGCGUAAUUGACAUCUGAUUGAAG_AAGCAUCUGGCUGCUUGCCAGGUCC___GC_UACGUGCCACCUGGCUAAUCACACAGCCAGGCACGACAGGUGUGGA____UAGAGAUGUGUAG ...........(((((((............(((((((.....)))))))..........((((((...(((((........))))))))))).))))))).................... (-24.61 = -26.75 + 2.14)

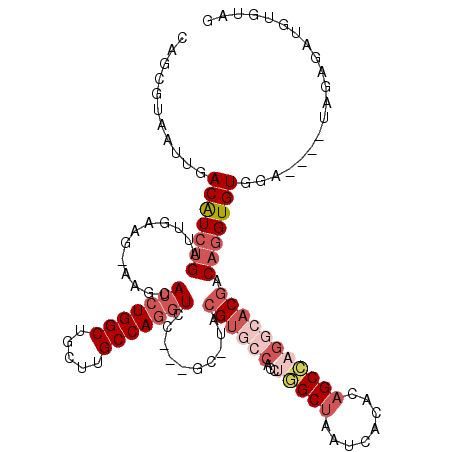
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:03 2006