
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,260,108 – 3,260,256 |
| Length | 148 |
| Max. P | 0.971887 |

| Location | 3,260,108 – 3,260,216 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.64 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.94 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3260108 108 - 22407834 UGAAUGCAGAUCGUUAGUGAGUUUUAUGCCAUCCUUUGGCA-AACUGCAAUCGAGAAAC-CUUGAUUGCACCACC-ACACUAUU-CAAGGCUUAAAGCUUAAU--------AUCCCCGAC ..........(((.(((((((((...(((((.....)))))-))))(((((((((....-)))))))))......-.)))))..-..((((.....))))...--------.....))). ( -29.40) >DroSec_CAF1 86699 108 - 1 UGACUGCAGAUCGUUAGUGAGUUUUAUGCCAUCCUUUGGCA-AACUGCAAUCGAGAAGU-CCUGAUUGCACCACC-ACACUAUC-CAAGGCUUAAAGCUUGAU--------AUCCCCGAC ..........(((.(((((((((...(((((.....)))))-))))(((((((.(....-).)))))))......-.)))))..-..((((.....))))...--------.....))). ( -25.50) >DroSim_CAF1 89817 108 - 1 UGACUGCAGAUCGUUAGUGAGUUUUAUGCCAUCCUUUGGCA-AACUGCAAUCGAGAAGC-CCUGAUUGCACCACC-ACACUAUC-CAAGGCUUAAAGCUUGAU--------AUCCCCUAC .........(((..(((((((((...(((((.....)))))-))))(((((((......-..)))))))......-.)))))..-..((((.....)))))))--------......... ( -24.80) >DroEre_CAF1 83549 105 - 1 UGCCUGCAGAUCGUUAGUGAGUUUUAUGCCAUCCUUUGGCAAAACUGCAAUCGAGAAA--CCUGAUUGCACUA----CACCAUA-CCAGGCUUAAAGCAUGAA--------UUCCCUGGC .(((((...((.((((((.(((((..(((((.....))))))))))((((((.((...--.))))))))))))----.)).)).-.)))))............--------......... ( -28.20) >DroYak_CAF1 82950 119 - 1 UGACUGCAGAUCGUUAGUGAGUUCUGUGCCAUCCUUCGGCA-AACUGCAAUCGAGAAUCCCCUGAUUGCACUACUAACACUAUCACAUGGCUUAAAGCUUGAUUUUCCGAUUUCCCUCAC (((..(.((((((((((((((((...((((.......))))-))))((((((.((......))))))))..)))))))...((((...(((.....))))))).....))))))..))). ( -28.50) >consensus UGACUGCAGAUCGUUAGUGAGUUUUAUGCCAUCCUUUGGCA_AACUGCAAUCGAGAAAC_CCUGAUUGCACCACC_ACACUAUC_CAAGGCUUAAAGCUUGAU________AUCCCCGAC .....((.......(((((((((...(((((.....))))).))))((((((.((......))))))))........))))).......))............................. (-22.70 = -22.94 + 0.24)

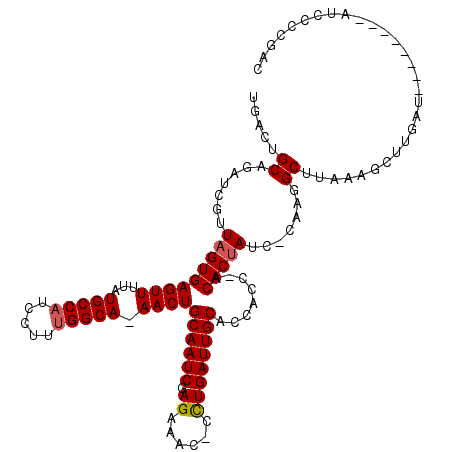
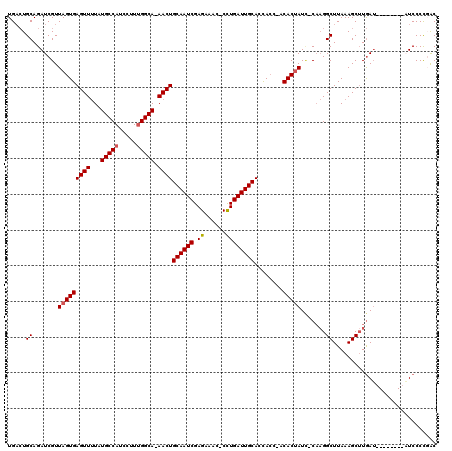
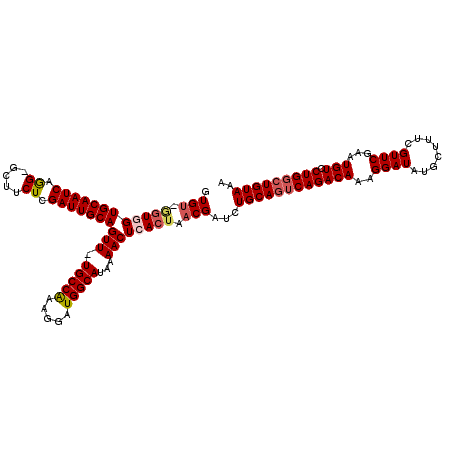
| Location | 3,260,139 – 3,260,256 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -31.84 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3260139 117 + 22407834 GUGU-GGUGGUGCAAUCAAG-GUUUCUCGAUUGCAGUU-UGCCAAAGGAUGGCAUAAAACUCACUAACGAUCUGCAUUCAGACAAAGGAUAUGCUUUCGUUCGAAUGUCCUGGCUGUAAA ...(-((((((((((((.((-....)).)))))))...-(((((.....)))))......))))))(((..(.((((((..((((((......)))).))..))))))...)..)))... ( -32.00) >DroSec_CAF1 86730 117 + 1 GUGU-GGUGGUGCAAUCAGG-ACUUCUCGAUUGCAGUU-UGCCAAAGGAUGGCAUAAAACUCACUAACGAUCUGCAGUCAGACAAAGGAUAUGCUUUCGUUCGAAUGUCCUGGCUGUAAA ((.(-((((((((((((..(-....)..)))))))...-(((((.....)))))......))))))))....((((((((((((..((((........))))...))).))))))))).. ( -37.70) >DroSim_CAF1 89848 117 + 1 GUGU-GGUGGUGCAAUCAGG-GCUUCUCGAUUGCAGUU-UGCCAAAGGAUGGCAUAAAACUCACUAACGAUCUGCAGUCAGACAAAGGAUAUGCUUUCGUUCGAAUGUCCUGGCUGUAAA ((.(-((((((((((((..(-....)..)))))))...-(((((.....)))))......))))))))....((((((((((((..((((........))))...))).))))))))).. ( -36.80) >DroEre_CAF1 83580 114 + 1 GUG----UAGUGCAAUCAGG--UUUCUCGAUUGCAGUUUUGCCAAAGGAUGGCAUAAAACUCACUAACGAUCUGCAGGCAGACAAAGGAUAUGCUCUCGUUCGAAUGUCCUGGCUGUAAA ((.----((((((((((((.--...)).))))))((((((((((.....)))))..))))).))))))....(((((.((((((..((((........))))...))).))).))))).. ( -32.20) >DroYak_CAF1 82990 119 + 1 GUGUUAGUAGUGCAAUCAGGGGAUUCUCGAUUGCAGUU-UGCCGAAGGAUGGCACAGAACUCACUAACGAUCUGCAGUCAGACAAAGGAUAUGCUCUCGUUCGAAUGUCCUGGCUGUAAA .((((((((((((((((.(((....)))))))))....-(((((.....)))))....))).)))))))...((((((((((((..((((........))))...))).))))))))).. ( -38.50) >consensus GUGU_GGUGGUGCAAUCAGG_GCUUCUCGAUUGCAGUU_UGCCAAAGGAUGGCAUAAAACUCACUAACGAUCUGCAGUCAGACAAAGGAUAUGCUUUCGUUCGAAUGUCCUGGCUGUAAA .(((.((((((((((((.((.....)).)))))))(((.(((((.....)))))...)))))))).)))...((((((((((((..((((........))))...))).))))))))).. (-31.84 = -32.80 + 0.96)


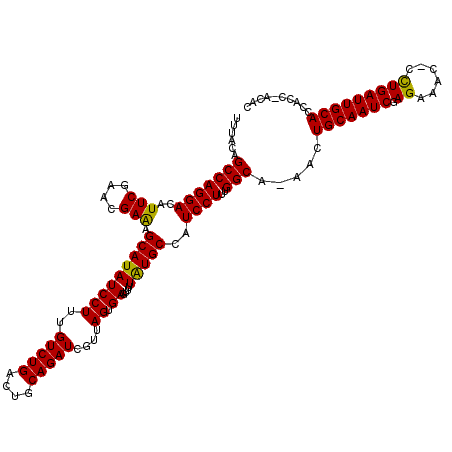
| Location | 3,260,139 – 3,260,256 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -26.16 |
| Energy contribution | -25.60 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3260139 117 - 22407834 UUUACAGCCAGGACAUUCGAACGAAAGCAUAUCCUUUGUCUGAAUGCAGAUCGUUAGUGAGUUUUAUGCCAUCCUUUGGCA-AACUGCAAUCGAGAAAC-CUUGAUUGCACCACC-ACAC ......((..(((((...((.(....)....))...)))))....)).........(((((((...(((((.....)))))-))))(((((((((....-))))))))).....)-)).. ( -30.80) >DroSec_CAF1 86730 117 - 1 UUUACAGCCAGGACAUUCGAACGAAAGCAUAUCCUUUGUCUGACUGCAGAUCGUUAGUGAGUUUUAUGCCAUCCUUUGGCA-AACUGCAAUCGAGAAGU-CCUGAUUGCACCACC-ACAC ....((..((((((.(((...(((..(((....(((...(((((........))))).))).....(((((.....)))))-...)))..))).)))))-))))..)).......-.... ( -30.30) >DroSim_CAF1 89848 117 - 1 UUUACAGCCAGGACAUUCGAACGAAAGCAUAUCCUUUGUCUGACUGCAGAUCGUUAGUGAGUUUUAUGCCAUCCUUUGGCA-AACUGCAAUCGAGAAGC-CCUGAUUGCACCACC-ACAC ......(((((((..(((....))).(((((..(((...(((((........))))).)))...)))))..)))..)))).-...((((((((......-..)))))))).....-.... ( -27.90) >DroEre_CAF1 83580 114 - 1 UUUACAGCCAGGACAUUCGAACGAGAGCAUAUCCUUUGUCUGCCUGCAGAUCGUUAGUGAGUUUUAUGCCAUCCUUUGGCAAAACUGCAAUCGAGAAA--CCUGAUUGCACUA----CAC ......((.(((.((..((((.((.......)).))))..))))))).....((.(((.(((((..(((((.....))))))))))((((((.((...--.))))))))))))----).. ( -29.10) >DroYak_CAF1 82990 119 - 1 UUUACAGCCAGGACAUUCGAACGAGAGCAUAUCCUUUGUCUGACUGCAGAUCGUUAGUGAGUUCUGUGCCAUCCUUCGGCA-AACUGCAAUCGAGAAUCCCCUGAUUGCACUACUAACAC ....(((...(((((...((.(....)....))...)))))..)))......(((((((((((...((((.......))))-))))((((((.((......))))))))..))))))).. ( -28.60) >consensus UUUACAGCCAGGACAUUCGAACGAAAGCAUAUCCUUUGUCUGACUGCAGAUCGUUAGUGAGUUUUAUGCCAUCCUUUGGCA_AACUGCAAUCGAGAAAC_CCUGAUUGCACCACC_ACAC ......(((((((..(((....))).(((((((((..(((((....)))))....)).))....)))))..))))..))).....(((((((.((......))))))))).......... (-26.16 = -25.60 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:00 2006